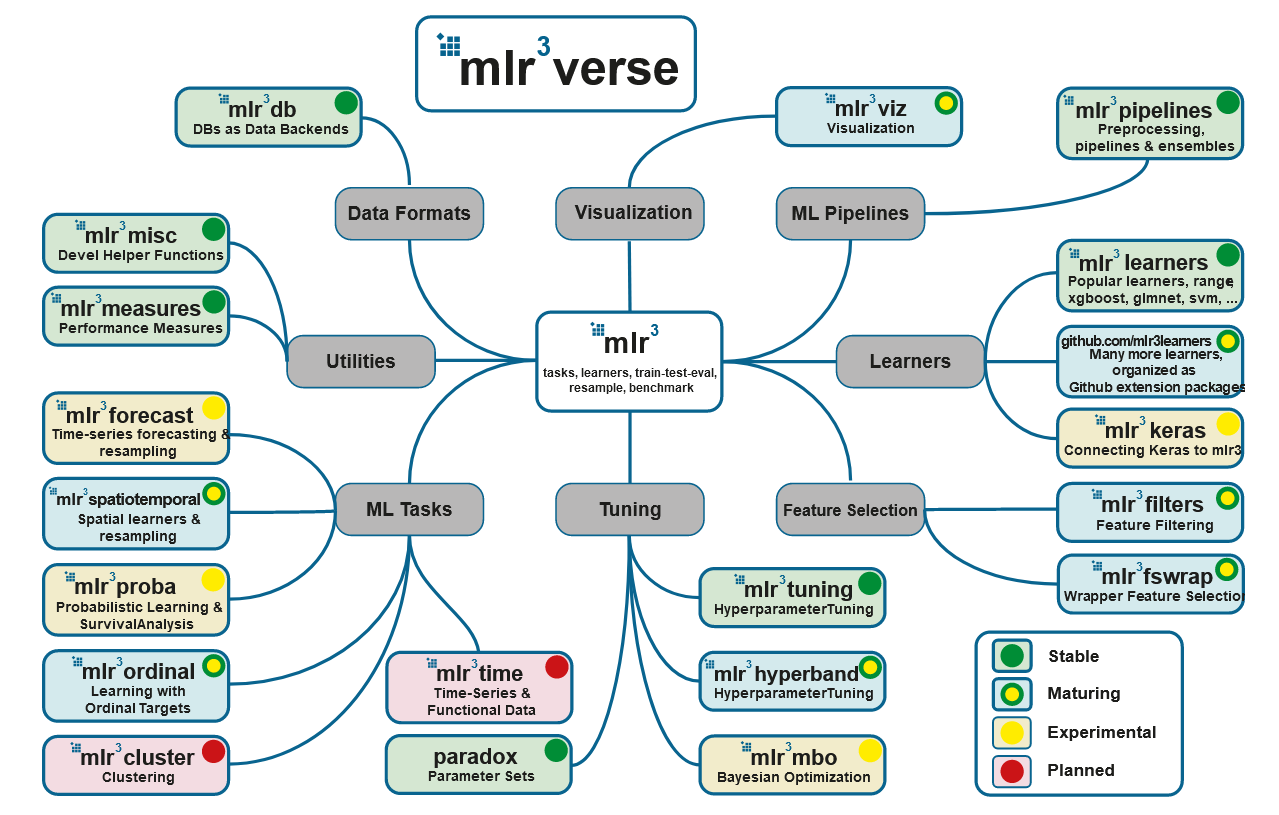
Fuente: https://mlr3book.mlr-org.com/
Hola Habr!En esta publicación, consideraremos el enfoque más reflexivo para el aprendizaje automático en el lenguaje R hoy: el paquete mlr3 y el ecosistema que lo rodea. Este enfoque se basa en la OOP "normal" utilizando clases R6 y en representar todas las operaciones con datos y modelos en forma de un gráfico de cálculo. Esto le permite crear tuberías optimizadas y flexibles para las tareas de aprendizaje automático, pero al principio puede parecer complicado y confuso. A continuación intentaremos aportar algo de claridad y motivarnos para usar mlr3 en sus proyectos.Contenido:
- Un poco de historia y comparación con soluciones de la competencia.
- Detalles técnicos: clases R6 y paquete data.table
- Los componentes principales de la tubería ML en mlr3
- Establecer hiperparámetros
- Descripción general del ecosistema mlr3
- Tuberías y gráfico de cálculos.
1.
caret — ,
caret R ( CRAN 2007 ). 2013 Applied Predictive Modeling, .
:
- ( - );
- (-), , , ;
- , caret- - ;
- — , xgboost
nrounds, max_depth, eta, gamma, colsample_bytree, min_child_weight subsample.
:
- — , . ;
- : , . recipes , ;
- - (nested resampling), caretEnsemble.
tidyverse strikes back
tidymodels, recipes ( «» , ), rsample ( ) tune ( ).
:
- «» , ;
- , embed textrecipes;
- , . ( tune);
- workflows «» .
:
- , tune . «» , ,
apply/map-; - . , 200 ;
- - .
mlr3 vs
mlr3 mlr , caret tidymodels. mlr , mlr3.
:
- R6-, data.table;
- . , ;
- API ,
learner — ; - .
:
2. : R6- data.table
mlr3 «» , R6-. R6- , . Advanced R, , .
R6- R6Class():
library(R6)
Accumulator <- R6Class("Accumulator", list(
sum = 0,
add = function(x = 1) {
self$sum <- self$sum + x
invisible(self)
})
)
— "Accumulator".
new(), (, , ) :
x <- Accumulator$new()
, , :
x$add(4)
x$sum
#> [1] 4
R6- :
y1 <- Accumulator$new()
y2 <- y1
y1$add(10)
c(y1 = y1$sum, y2 = y2$sum)
#> y1 y2
#> 10 10
clone() ( clone(deep = TRUE) ):
y1 <- Accumulator$new()
y2 <- y1$clone()
y1$add(10)
c(y1 = y1$sum, y2 = y2$sum)
#> y1 y2
#> 10 0
, R6 mlr3.
data.table ( , data.table data.table: R). - data.table, := . , , 2 , . , , .
3. ML- mlr3

: https://mlr3book.mlr-org.com/
mlr3 :
library(mlr3)
#
task <- TaskClassif$new(id = "iris",
backend = iris,
target = "Species")
task
# <TaskClassif:iris> (150 x 5)
# * Target: Species
# * Properties: multiclass
# * Features (4):
# - dbl (4): Petal.Length, Petal.Width, Sepal.Length, Sepal.Width
#
# learner_rpart <- mlr_learners$get("classif.rpart")
learner_rpart <- lrn("classif.rpart",
predict_type = "prob",
minsplit = 50)
learner_rpart
# <LearnerClassifRpart:classif.rpart>
# * Model: -
# * Parameters: xval=0, minsplit=50
# * Packages: rpart
# * Predict Type: prob
# * Feature types: logical, integer, numeric, factor, ordered
# * Properties: importance, missings, multiclass, selected_features, twoclass, weights
#
learner_rpart$param_set
# ParamSet:
# id class lower upper levels default value
# 1: minsplit ParamInt 1 Inf 20 50
# 2: minbucket ParamInt 1 Inf <NoDefault>
# 3: cp ParamDbl 0 1 0.01
# 4: maxcompete ParamInt 0 Inf 4
# 5: maxsurrogate ParamInt 0 Inf 5
# 6: maxdepth ParamInt 1 30 30
# 7: usesurrogate ParamInt 0 2 2
# 8: surrogatestyle ParamInt 0 1 0
# 9: xval ParamInt 0 Inf 10 0
#
learner_rpart$train(task, row_ids = 1:120)
learner_rpart$model
# n= 120
#
# node), split, n, loss, yval, (yprob)
# * denotes terminal node
#
# 1) root 120 70 setosa (0.41666667 0.41666667 0.16666667)
# 2) Petal.Length< 2.45 50 0 setosa (1.00000000 0.00000000 0.00000000) *
# 3) Petal.Length>=2.45 70 20 versicolor (0.00000000 0.71428571 0.28571429)
# 6) Petal.Length< 4.95 49 1 versicolor (0.00000000 0.97959184 0.02040816) *
# 7) Petal.Length>=4.95 21 2 virginica (0.00000000 0.09523810 0.90476190) *
: (task) (learner).
(TaskClassif , TaskRegr ..) new(). id, backend target; positive. : mlr_tasks$get("iris") tsk("iris").
mlr_learners get() train(), task , . : lrn("classif.rpart", predict_type = "prob", minsplit = 50). (predict_type = "prob") (minsplit = 50). : learner_rpart$predict_type <- "prob", learner_rpart$param_set$values$minsplit = 50.
predict_newdata():
#
preds <- learner_rpart$predict_newdata(newdata = iris[121:150, ])
preds
# <PredictionClassif> for 30 observations:
# row_id truth response prob.setosa prob.versicolor prob.virginica
# 1 virginica virginica 0 0.0952381 0.90476190
# 2 virginica versicolor 0 0.9795918 0.02040816
# 3 virginica virginica 0 0.0952381 0.90476190
# ---
# 28 virginica virginica 0 0.0952381 0.90476190
# 29 virginica virginica 0 0.0952381 0.90476190
# 30 virginica virginica 0 0.0952381 0.90476190
- 5 :
cv10 <- rsmp("cv", folds = 5)
resample_results <- resample(task, learner_rpart, cv10)
# INFO [09:37:05.993] Applying learner 'classif.rpart' on task 'iris' (iter 1/5)
# INFO [09:37:06.018] Applying learner 'classif.rpart' on task 'iris' (iter 2/5)
# INFO [09:37:06.042] Applying learner 'classif.rpart' on task 'iris' (iter 3/5)
# INFO [09:37:06.074] Applying learner 'classif.rpart' on task 'iris' (iter 4/5)
# INFO [09:37:06.098] Applying learner 'classif.rpart' on task 'iris' (iter 5/5)
resample_results
# <ResampleResult> of 5 iterations
# * Task: iris
# * Learner: classif.rpart
# * Warnings: 0 in 0 iterations
# * Errors: 0 in 0 iterations
# (-):
as.data.table(mlr_resamplings)
# key params iters
# 1: bootstrap repeats,ratio 30
# 2: custom 0
# 3: cv folds 10
# 4: holdout ratio 1
# 5: repeated_cv repeats,folds 100
# 6: subsampling repeats,ratio 30
. score() resample_results, — accuracy "classif.acc" classification error "classif.ce". , get(): mlr_measures$get("classif.ce"). msrs():
resample_results$score(msrs(c("classif.acc", "classif.ce")))[, 5:10]
#
# resampling resampling_id iteration prediction classif.acc classif.ce
# 1: <ResamplingCV> cv 1 <list> 0.8666667 0.13333333
# 2: <ResamplingCV> cv 2 <list> 0.9666667 0.03333333
# 3: <ResamplingCV> cv 3 <list> 0.9333333 0.06666667
# 4: <ResamplingCV> cv 4 <list> 0.9666667 0.03333333
# 5: <ResamplingCV> cv 5 <list> 0.9333333 0.06666667
4.
— . , .
. paradox:
library("paradox")
searchspace <- ParamSet$new(list(
ParamDbl$new("cp", lower = 0.001, upper = 0.1),
ParamInt$new("minsplit", lower = 1, upper = 10)
))
searchspace
# ParamSet:
# id class lower upper levels default value
# 1: cp ParamDbl 0.001 0.1 <NoDefault>
# 2: minsplit ParamInt 1.000 10.0 <NoDefault>
ParamSet, cp minsplit; rpart .
, searchspace . tune() Tuner. . resolution, , param_resolutions, . , , .
generate_design_grid() , :
generate_design_grid(searchspace,
param_resolutions = c("cp" = 2, "minsplit" = 3))
# <Design> with 6 rows:
# cp minsplit
# 1: 0.001 1
# 2: 0.001 5
# 3: 0.001 10
# 4: 0.100 1
# 5: 0.100 5
# 6: 0.100 10
: generate_design_random() generate_design_lhs() .
, . Terminator-, , ( ), . mlr3tuning:
library("mlr3tuning")
evals20 <- term("evals", n_evals = 20)
evals20
# <TerminatorEvals>
# * Parameters: n_evals=20
#
as.data.table(mlr_terminators)
# key
# 1: clock_time
# 2: combo
# 3: evals
# 4: model_time
# 5: none
# 6: perf_reached
# 7: stagnation
TuningInstance:
tuning_instance <- TuningInstance$new(
task = TaskClassif$new(id = "iris",
backend = iris,
target = "Species"),
learner = lrn("classif.rpart",
predict_type = "prob"),
resampling = rsmp("cv", folds = 5),
measures = msr("classif.ce"),
param_set = ParamSet$new(
list(ParamDbl$new("cp", lower = 0.001, upper = 0.1),
ParamInt$new("minsplit", lower = 1, upper = 10)
)
),
terminator = term("evals", n_evals = 20)
)
tuning_instance
# <TuningInstance>
# * State: Not tuned
# * Task: <TaskClassif:iris>
# * Learner: <LearnerClassifRpart:classif.rpart>
# * Measures: classif.ce
# * Resampling: <ResamplingCV>
# * Terminator: <TerminatorEvals>
# * bm_args: list()
# * n_evals: 0
# ParamSet:
# id class lower upper levels default value
# 1: cp ParamDbl 0.001 0.1 <NoDefault>
# 2: minsplit ParamInt 1.000 10.0 <NoDefault>
— Tuner, :
tuner <- tnr("grid_search",
resolution = 5,
batch_size = 2)
#
# as.data.table(mlr_tuners)
# key
# 1: design_points
# 2: gensa
# 3: grid_search
# 4: random_search
resolution = 5, 25 . 20 , terminator = term("evals", n_evals = 20). batch_size — , . mlr3 — , .
tnr("design_points"): , ( — , mlr3 ).
, :
result <- tuner$tune(tuning_instance)
result
# NULL
, result . , tuner$tune() tuning_instance:
tuning_instance$result
# $tune_x
# $tune_x$cp
# [1] 0.001
#
# $tune_x$minsplit
# [1] 5
#
#
# $params
# $params$xval
# [1] 0
#
# $params$cp
# [1] 0.001
#
# $params$minsplit
# [1] 5
#
#
# $perf
# classif.ce
# 0.04
result <- tuning_instance$archive(unnest = "params")
result[order(classif.ce), c("cp", "minsplit", "classif.ce")]
# cp minsplit classif.ce
# 1: 0.00100 5 0.04000000
# 2: 0.00100 3 0.04000000
# 3: 0.00100 8 0.04000000
# 4: 0.00100 1 0.04000000
# 5: 0.00100 10 0.04666667
# 6: 0.02575 10 0.06000000
# 7: 0.07525 5 0.06000000
# 8: 0.02575 8 0.06000000
# 9: 0.02575 3 0.06000000
# 10: 0.05050 1 0.06000000
# 11: 0.07525 3 0.06000000
# 12: 0.07525 1 0.06000000
# 13: 0.05050 3 0.06000000
# 14: 0.02575 5 0.06000000
# 15: 0.05050 5 0.06000000
# 16: 0.05050 8 0.06000000
# 17: 0.10000 3 0.06000000
# 18: 0.10000 8 0.06000000
# 19: 0.05050 10 0.06000000
# 20: 0.10000 1 0.06000000
library(ggplot2)
ggplot(result,
aes(x = cp, y = classif.ce, color = as.factor(minsplit))) +
geom_line() +
geom_point(size = 3)

, tune():
Tuner ( batch_size);Learner Task . ResampleResult ( BenchmarkResult);Terminator , . , 1, , ;- ;
- , ( , ,
msr("classif.ce", aggregator = "median").
tuning_instance$bmr, BenchmarkResult, score() as.data.table(tuning_instance$bmr). , , ResampleResult tuning_instance$archive():
tuning_instance$archive()[1, resample_result][[1]]$score()[, 4:9]
# learner_id resampling resampling_id iteration prediction classif.ce
# 1: classif.rpart <ResamplingCV> cv 1 <list> 0.06666667
# 2: classif.rpart <ResamplingCV> cv 2 <list> 0.16666667
# 3: classif.rpart <ResamplingCV> cv 3 <list> 0.03333333
# 4: classif.rpart <ResamplingCV> cv 4 <list> 0.03333333
# 5: classif.rpart <ResamplingCV> cv 5 <list> 0.00000000
, :
res <- tuning_instance$archive(unnest = "params")
res[, ce_resemples := lapply(resample_result, function(x) x$score()[, classif.ce])]
ce_resemples <- res[, .(ce_resemples = unlist(ce_resemples)), by = nr]
res[ce_resemples, on = "nr"]
5. mlr3
: mlr3, mlr3tuning paradox. , - mlr3verse:
# install.packages("mlr3verse")
library(mlr3verse)
## Loading required package: mlr3
## Loading required package: mlr3db
## Loading required package: mlr3filters
## Loading required package: mlr3learners
## Loading required package: mlr3pipelines
## Loading required package: mlr3tuning
## Loading required package: mlr3viz
## Loading required package: paradox
- mlr3db dbplyr data.table.
- mlr3filters , ( ).
- mlr3learners (
regr.glmnet, regr.kknn, regr.km, regr.lm, regr.ranger, regr.svm, regr.xgboost) (classif.glmnet, classif.kknn, classif.lda, classif.log_reg, classif.multinom, classif.naive_bayes, classif.qda, classif.ranger, classif.svm, classif.xgboost). . - mlr3pipelines (pipelines), . , , CRAN, :
remotes::install_github("https://github.com/mlr-org/mlr3pipelines"). - mlr3tuning .
- mlr3viz , .
- mlr3measures — ~40 . mlr3verse , .
, .
6.
(pipelines) , , .
— , , — . PipeOpLearner(), — PipeOpFilter(), — PipeOp(). ( po()) :
pca <- po("pca")
filter <- po("filter",
filter = mlr3filters::flt("variance"),
filter.frac = 0.5)
learner_po <- po("learner",
learner = lrn("classif.rpart"))
%>>%:
graph <- pca %>>% filter %>>% learner_po
graph$plot()
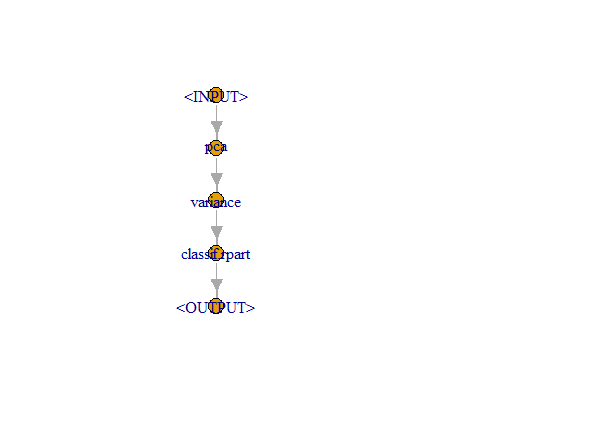
. , :
gr <- Graph$new()$
add_pipeop(mlr_pipeops$get("copy", outnum = 2))$
add_pipeop(mlr_pipeops$get("scale"))$
add_pipeop(mlr_pipeops$get("pca"))$
add_pipeop(mlr_pipeops$get("featureunion", innum = 2))
gr$
add_edge("copy", "scale", src_channel = 1)$
add_edge("copy", "pca", src_channel = "output2")$
add_edge("scale", "featureunion", dst_channel = 1)$
add_edge("pca", "featureunion", dst_channel = 2)
gr$plot(html = FALSE)

, (po("learner", learner = lrn("classif.rpart"))). , :
glrn <- GraphLearner$new(graph)
glrn
# <GraphLearner:pca.variance.classif.rpart>
# * Model: -
# * Parameters: variance.filter.frac=0.5, variance.na.rm=TRUE, classif.rpart.xval=0
# * Packages: -
# * Predict Type: response
# * Feature types: logical, integer, numeric, character, factor, ordered, POSIXct
# * Properties: importance, missings, multiclass, oob_error, selected_features, twoclass,
# weights
GraphLearner Learner. , Learner-, :
resample(tsk("iris"), glrn, rsmp("cv"))
# INFO [17:17:00.358] Applying learner 'pca.variance.classif.rpart' on task 'iris' (iter 1/10)
# INFO [17:17:00.615] Applying learner 'pca.variance.classif.rpart' on task 'iris' (iter 2/10)
# INFO [17:17:00.881] Applying learner 'pca.variance.classif.rpart' on task 'iris' (iter 3/10)
# INFO [17:17:01.087] Applying learner 'pca.variance.classif.rpart' on task 'iris' (iter 4/10)
# INFO [17:17:01.303] Applying learner 'pca.variance.classif.rpart' on task 'iris' (iter 5/10)
# INFO [17:17:01.518] Applying learner 'pca.variance.classif.rpart' on task 'iris' (iter 6/10)
# INFO [17:17:01.716] Applying learner 'pca.variance.classif.rpart' on task 'iris' (iter 7/10)
# INFO [17:17:01.927] Applying learner 'pca.variance.classif.rpart' on task 'iris' (iter 8/10)
# INFO [17:17:02.129] Applying learner 'pca.variance.classif.rpart' on task 'iris' (iter 9/10)
# INFO [17:17:02.337] Applying learner 'pca.variance.classif.rpart' on task 'iris' (iter 10/10)
# <ResampleResult> of 10 iterations
# * Task: iris
# * Learner: pca.variance.classif.rpart
# * Warnings: 0 in 0 iterations
# * Errors: 0 in 0 iterations
, issue How to deal with different preprocessing steps as hyperparameters:
gr <- pipeline_branch(list(pca = po("pca"), nothing = po("nop")))
gr$plot()

caret tidymodels !
, mlr3. mlr3 book .