La detección de anomalías es una tarea interesante de aprendizaje automático. No hay una forma específica de resolverlo, ya que cada conjunto de datos tiene sus propias características. Pero al mismo tiempo, hay varios enfoques que ayudan a tener éxito. Quiero hablar sobre uno de estos enfoques: autoencoders.
¿Qué conjunto de datos elegir?
El problema más apremiante en la vida de cualquier científico de datos. Para simplificar la historia, usaré un conjunto de datos simple en la estructura, que generaremos aquí.
import os
import numpy as np
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
from matplotlib.colors import Normalize
def gen_normal_distribution(mu, sigma, size, range=(0, 1), max_val=1):
bins = np.linspace(*range, size)
result = 1 / (sigma * np.sqrt(2*np.pi)) * np.exp(-(bins - mu)**2 / (2*sigma**2))
cur_max_val = result.max()
k = max_val / cur_max_val
result *= k
return result
Considere un ejemplo de una función. Cree una distribución normal con μ = 0.3 y σ = 0.05:
dist = gen_normal_distribution(0.3, 0.05, 256, max_val=1)
print(dist.max())
>>> 1.0
plt.plot(np.linspace(0, 1, 256), dist)
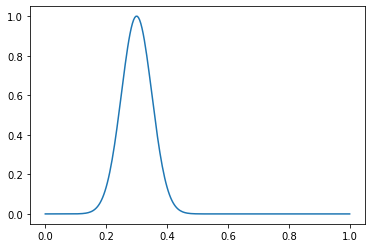
Declarar los parámetros de nuestro conjunto de datos:
in_distribution_size = 2000
out_distribution_size = 200
val_size = 100
sample_size = 256
random_generator = np.random.RandomState(seed=42)
Y las funciones para generar ejemplos son normales y anormales. Las distribuciones con un máximo se considerarán normales, anormales, con dos:
def generate_in_samples(size, sample_size):
global random_generator
in_samples = np.zeros((size, sample_size))
in_mus = random_generator.uniform(0.1, 0.9, size)
in_sigmas = random_generator.uniform(0.05, 0.5, size)
for i in range(size):
in_samples[i] = gen_normal_distribution(in_mus[i], in_sigmas[i], sample_size, max_val=1)
return in_samples
def generate_out_samples(size, sample_size):
global random_generator
out_samples = generate_in_samples(size, sample_size)
out_additional_mus = random_generator.uniform(0.1, 0.9, size)
out_additional_sigmas = random_generator.uniform(0.01, 0.05, size)
for i in range(size):
anomaly = gen_normal_distribution(out_additional_mus[i], out_additional_sigmas[i], sample_size, max_val=0.12)
out_samples[i] += anomaly
return out_samples
Así es como se ve un ejemplo normal:
in_samples = generate_in_samples(in_distribution_size, sample_size)
plt.plot(np.linspace(0, 1, sample_size), in_samples[42])

Y anormal, como este:
out_samples = generate_out_samples(out_distribution_size, sample_size)
plt.plot(np.linspace(0, 1, sample_size), out_samples[42])
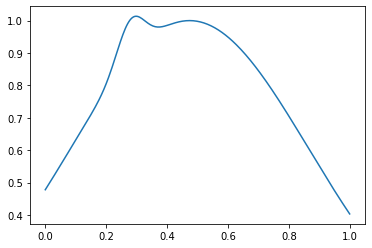
Crear matrices con etiquetas y etiquetas:
x = np.concatenate((in_samples, out_samples))
y = np.concatenate((np.zeros(in_distribution_size), np.ones(out_distribution_size)))
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, shuffle=True, random_state=42)
. , , 2 1 (). , ( ), :
x_val_out = generate_out_samples(val_size, sample_size)
x_val_in = generate_in_samples(val_size, sample_size)
x_val = np.concatenate((x_val_out, x_val_in))
y_val = np.concatenate((np.ones(val_size), np.zeros(val_size)))
, , Sklearn: SVM . , , .
from sklearn.metrics import classification_report
from sklearn.metrics import f1_score
One class SVM
from sklearn.svm import OneClassSVM
OneClassSVM nu — .
out_dist_part = out_distribution_size / (out_distribution_size + in_distribution_size)
svm = OneClassSVM(nu=out_dist_part)
svm.fit(x_train, y_train)
>>> OneClassSVM(cache_size=200, coef0=0.0, degree=3, gamma='scale', kernel='rbf',
max_iter=-1, nu=0.09090909090909091, shrinking=True, tol=0.001,
verbose=False)
:
svm_prediction = svm.predict(x_val)
svm_prediction[svm_prediction == 1] = 0
svm_prediction[svm_prediction == -1] = 1
sklearn — classification_report, Anomaly detection , precision recall, :
print(classification_report(y_val, svm_prediction))
>>> precision recall f1-score support
0.0 0.57 0.93 0.70 100
1.0 0.81 0.29 0.43 100
accuracy 0.61 200
macro avg 0.69 0.61 0.57 200
weighted avg 0.69 0.61 0.57 200
, . f1-score , .
Isolation forest
, , - ?
from sklearn.ensemble import IsolationForest
, . :
out_dist_part = out_distribution_size / (out_distribution_size + in_distribution_size)
iso_forest = IsolationForest(n_estimators=100, contamination=out_dist_part, max_features=100, n_jobs=-1)
iso_forest.fit(x_train)
>>> IsolationForest(behaviour='deprecated', bootstrap=False,
contamination=0.09090909090909091, max_features=100,
max_samples='auto', n_estimators=100, n_jobs=-1,
random_state=None, verbose=0, warm_start=False)
Classification report? — Classification report!
iso_forest_prediction = iso_forest.predict(x_val)
iso_forest_prediction[iso_forest_prediction == 1] = 0
iso_forest_prediction[iso_forest_prediction == -1] = 1
print(classification_report(y_val, iso_forest_prediction))
>>> precision recall f1-score support
0.0 0.50 0.91 0.65 100
1.0 0.53 0.10 0.17 100
accuracy 0.51 200
macro avg 0.51 0.51 0.41 200
weighted avg 0.51 0.51 0.41 200
RandomForestClassifier
, - " ?" , :
from sklearn.ensemble import RandomForestClassifier
random_forest = RandomForestClassifier(n_estimators=100, max_features=100, n_jobs=-1)
random_forest.fit(x_train, y_train)
>>> RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=None, max_features=100,
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=100,
n_jobs=-1, oob_score=False, random_state=None, verbose=0,
warm_start=False)
random_forest_prediction = random_forest.predict(x_val)
print(classification_report(y_val, random_forest_prediction))
>>> precision recall f1-score support
0.0 0.57 0.99 0.72 100
1.0 0.96 0.25 0.40 100
accuracy 0.62 200
macro avg 0.77 0.62 0.56 200
weighted avg 0.77 0.62 0.56 200
Autoencoder
, : . .
, "" , . 2 : Encoder' Decoder', .
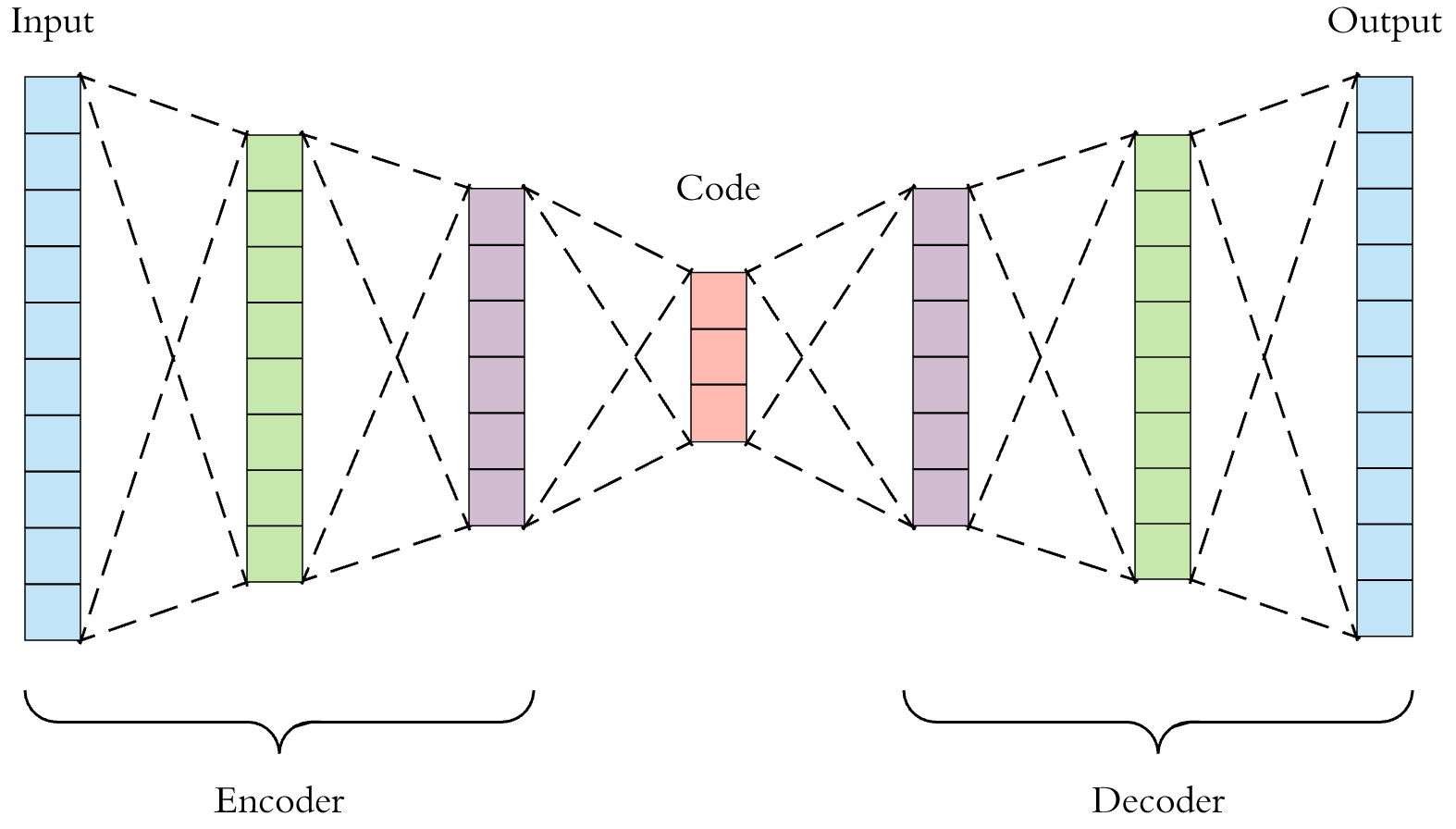
"" , , .
? , , , . , 9% 91% . , . , , : .
.
PyTorch, :
import torch
from torch import nn
from torch.utils.data import TensorDataset, DataLoader
from torch.optim import Adam
:
batch_size = 32
lr = 1e-3
. , , "" . , ( , learning rate, ) , , .
train_in_distribution = x_train[y_train == 0]
train_in_distribution = torch.tensor(train_in_distribution.astype(np.float32))
train_in_dataset = TensorDataset(train_in_distribution)
train_in_loader = DataLoader(train_in_dataset, batch_size=batch_size, shuffle=True)
test_dataset = TensorDataset(
torch.tensor(x_test.astype(np.float32)),
torch.tensor(y_test.astype(np.long))
)
test_loader = DataLoader(test_dataset, batch_size=batch_size, shuffle=False)
val_dataset = TensorDataset(torch.tensor(x_val.astype(np.float32)))
val_loader = DataLoader(val_dataset, batch_size=batch_size, shuffle=False)
. 4 , ( 2 : μ, — σ, ; 4 , ).
class Autoencoder(nn.Module):
def __init__(self, input_size):
super(Autoencoder, self).__init__()
self.encoder = nn.Sequential(
nn.Linear(input_size, 128),
nn.LeakyReLU(0.2),
nn.Linear(128, 64),
nn.LeakyReLU(0.2),
nn.Linear(64, 16),
nn.LeakyReLU(0.2),
nn.Linear(16, 4),
nn.LeakyReLU(0.2),
)
self.decoder = nn.Sequential(
nn.Linear(4, 16),
nn.LeakyReLU(0.2),
nn.Linear(16, 64),
nn.LeakyReLU(0.2),
nn.Linear(64, 128),
nn.LeakyReLU(0.2),
nn.Linear(128, 256),
nn.LeakyReLU(0.2),
)
def forward(self, x):
x = self.encoder(x)
x = self.decoder(x)
return x
model = Autoencoder(sample_size).cuda()
criterion = nn.MSELoss()
per_sample_criterion = nn.MSELoss(reduction="none")
optimizer = Adam(model.parameters(), lr=lr, weight_decay=1e-5)
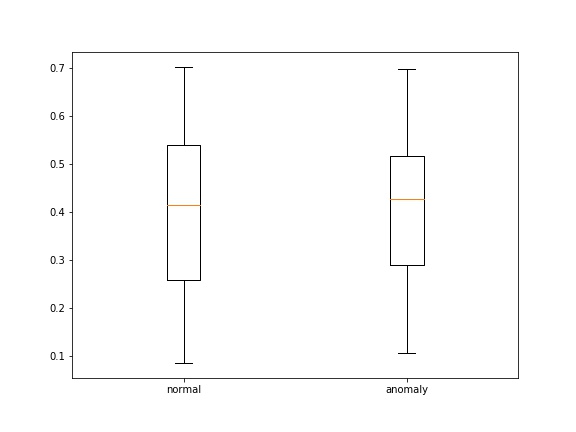
- loss' , , boxplot' :
def save_score_distribution(model, data_loader, criterion, save_to, figsize=(8, 6)):
losses = []
labels = []
for (x_batch, y_batch) in data_loader:
x_batch = x_batch.cuda()
output = model(x_batch)
loss = criterion(output, x_batch)
loss = torch.mean(loss, dim=1)
loss = loss.detach().cpu().numpy().flatten()
losses.append(loss)
labels.append(y_batch.detach().cpu().numpy().flatten())
losses = np.concatenate(losses)
labels = np.concatenate(labels)
losses_0 = losses[labels == 0]
losses_1 = losses[labels == 1]
fig, ax = plt.subplots(1, figsize=figsize)
ax.boxplot([losses_0, losses_1])
ax.set_xticklabels(['normal', 'anomaly'])
plt.savefig(save_to)
plt.close(fig)
:
:
experiment_path = "ood_detection"
!rm -rf $experiment_path
os.makedirs(experiment_path, exist_ok=True)
epochs = 100
for epoch in range(epochs):
running_loss = 0
for (x_batch, ) in train_in_loader:
x_batch = x_batch.cuda()
output = model(x_batch)
loss = criterion(output, x_batch)
optimizer.zero_grad()
loss.backward()
optimizer.step()
running_loss += loss.item()
print("epoch [{}/{}], train loss:{:.4f}".format(epoch+1, epochs, running_loss))
plot_path = os.path.join(experiment_path, "{}.jpg".format(epoch+1))
save_score_distribution(model, test_loader, per_sample_criterion, plot_path)
>>>
epoch [1/100], train loss:8.5728
epoch [2/100], train loss:4.2405
epoch [3/100], train loss:4.0852
epoch [4/100], train loss:1.7578
epoch [5/100], train loss:0.8543
...
epoch [96/100], train loss:0.0147
epoch [97/100], train loss:0.0154
epoch [98/100], train loss:0.0117
epoch [99/100], train loss:0.0105
epoch [100/100], train loss:0.0097

50

100
, . , :
def get_prediction(model, x):
global batch_size
dataset = TensorDataset(torch.tensor(x.astype(np.float32)))
data_loader = DataLoader(dataset, batch_size=batch_size, shuffle=False)
predictions = []
for batch in data_loader:
x_batch = batch[0].cuda()
pred = model(x_batch)
predictions.append(pred.detach().cpu().numpy())
predictions = np.concatenate(predictions)
return predictions
def compare_data(xs, sample_num, data_range=(0, 1), labels=None):
fig, axes = plt.subplots(len(xs))
sample_size = len(xs[0][sample_num])
for i in range(len(xs)):
axes[i].plot(np.linspace(*data_range, sample_size), xs[i][sample_num])
if labels:
for i, label in enumerate(labels):
axes[i].set_ylabel(label)
:
x_test_pred = get_prediction(model, x_test)
compare_data([x_test[y_test == 0], x_test_pred[y_test == 0]], 10, labels=["real", "encoded"])
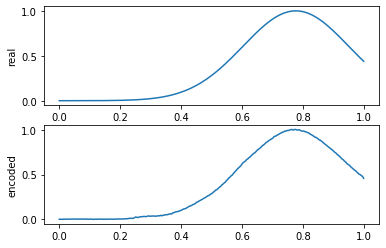
:

X? .
Difference score
— . , , , . .
def get_difference_score(model, x):
global batch_size
dataset = TensorDataset(torch.tensor(x.astype(np.float32)))
data_loader = DataLoader(dataset, batch_size=batch_size, shuffle=False)
predictions = []
for (x_batch, ) in data_loader:
x_batch = x_batch.cuda()
preds = model(x_batch)
predictions.append(preds.detach().cpu().numpy())
predictions = np.concatenate(predictions)
return (x - predictions)
from sklearn.ensemble import RandomForestClassifier
test_score = get_difference_score(model, x_test)
score_forest = RandomForestClassifier(max_features=100)
score_forest.fit(test_score, y_test)
>>> RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=None, max_features=100,
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=100,
n_jobs=None, oob_score=False, random_state=None,
verbose=0, warm_start=False)
, : 2 — . , 2 : , — difference_score, — . , , .
? . difference_score , ( ) , , . , , difference_score , ( ). .
:
val_score = get_difference_score(model, x_val)
prediction = score_forest.predict(val_score)
print(classification_report(y_val, prediction))
>>> precision recall f1-score support
0.0 0.76 1.00 0.87 100
1.0 1.00 0.69 0.82 100
accuracy 0.84 200
macro avg 0.88 0.84 0.84 200
weighted avg 0.88 0.84 0.84 200
. :
indices = np.arange(len(prediction))
wrong_indices = indices[(prediction == 0) & (y_val == 1)]
x_val_pred = get_prediction(model, x_val)
compare_data([x_val, x_val_pred], wrong_indices[0])
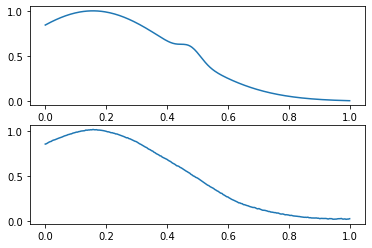
? :
plt.imshow(val_score[wrong_indices], norm=Normalize(0, 1, clip=True))

:
plt.imshow(val_score[(prediction == 1) & (y_val == 1)], norm=Normalize(0, 1, clip=True))

:
plt.imshow(val_score[(prediction == 0) & (y_val == 0)], norm=Normalize(0, 1, clip=True))

, : .
Difference histograms
. . — , "", .
, difference score
print("test score: [{}; {}]".format(test_score.min(), test_score.max()))
>>> test score: [-0.2260764424351479; 0.26339245919832344]
, :
def score_to_histograms(scores, bins=10, data_range=(-0.3, 0.3)):
result_histograms = np.zeros((len(scores), bins))
for i in range(len(scores)):
hist, bins = np.histogram(scores[i], bins=bins, range=data_range)
result_histograms[i] = hist
return result_histograms
test_histogram = score_to_histograms(test_score, bins=10, data_range=(-0.3, 0.3))
val_histogram = score_to_histograms(val_score, bins=10, data_range=(-0.3, 0.3))
plt.title("normal histogram")
plt.bar(np.linspace(-0.3, 0.3, 10), test_histogram[y_test == 0][0])

plt.title("anomaly histogram")
plt.bar(np.linspace(-0.3, 0.3, 10), test_histogram[y_test == 1][0])
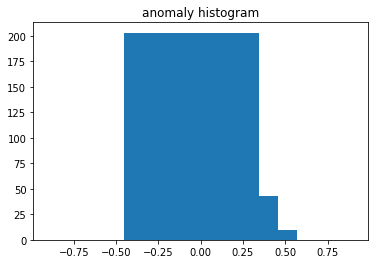
, "", .
histogram_forest = RandomForestClassifier(n_estimators=10)
histogram_forest.fit(test_histogram, y_test)
>>> RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=None, max_features='auto',
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=10,
n_jobs=None, oob_score=False, random_state=None,
verbose=0, warm_start=False)
val_prediction = histogram_forest.predict(val_histogram)
print(classification_report(y_val, val_prediction))
>>> precision recall f1-score support
0.0 0.83 0.99 0.90 100
1.0 0.99 0.80 0.88 100
accuracy 0.90 200
macro avg 0.91 0.90 0.89 200
weighted avg 0.91 0.90 0.89 200
— . , , , , . () .
— . ? VAE, ( 4 ) . . CVAE, - . , , , ..
GAN', (), . ( ).
, , .
Statistical parametric
- GMM — Gaussian mixture modelling + Akaike or Bayesian Information Criterion
- HMM — Hidden Markov models
- MRF — Markov random fields
- CRF — conditional random fields
Robust statistic
- minimum volume estimation
- PCA
- estimation maximisation (EM) + deterministic annealing
- K-means
Non-parametric statistics
- histogram analysis with density estimation on KNN
- local kernel models (Parzen windowing)
- vector of feature matching with similarity distance (between train and test)
- wavelets + MMRF
- histogram-based measures features
- texture features
- shape features
- features from VGG-16
- HOG
Neural networks
- self organisation maps (SOM) or Kohonen's
- Radial Basis Functions (RBF) (Minhas, 2005)
- LearningVector Quantisation (LVQ)
- ProbabilisticNeural Networks (PNN)
- Hopfieldnetworks
- SupportVector Machines (SVM)
- AdaptiveResonance Theory (ART)
- Relevance vector machine (RVM)
, Data science' . Computer vision, . ( — !) — FARADAY Lab. — , .
c:
