Introduction
. . . , , . ? ? , . : , , , . , , . , , , , . Johns Hopkins University.
COVID-19 — , SARS-CoV-2 (2019-nCoV). — , /, .
, , .
: |
, : , , , , , ( , ), . , , , , . , . , 2.4. , , , , — . , ( 15% ), , , ; .
, , , . . :
- ,
- .
:
- , .
- , ( ). , .
- , , . .
- - ( ) , .
— , - . COVID-19 1 14 .
. — . . , 0.35 ( , 2.4 , 1 14) 0.135 . , . .
Python
: . , , (Total cases), (New cases) (Infected), , ( - , ).
import numpy as np
import matplotlib.pyplot as plt
COUNTRY = "Italy"
DAYS_OF_SIMULATION = 366
COEF_BASE = 0.35
COEF_QUARANTINE = 0.135
DAY_QUARANTINE = 74
INCUBATION_PERIOD = 15
np.random.seed(0)
def get_coef(day):
return COEF_BASE if day < DAY_QUARANTINE else COEF_QUARANTINE
if __name__ == "__main__":
days = np.arange(1, DAYS_OF_SIMULATION)
infected = np.random.randint(1, INCUBATION_PERIOD, 1)
infected_lst = []
new_cases_lst = []
new_cases_total_lst = []
for day in days:
coef = get_coef(day)
new_cases_idx = np.argwhere(infected == day).flatten()
new_cases_count = new_cases_idx.size
infected = np.delete(infected, new_cases_idx)
new_infected_count = np.random.poisson(coef, infected.size).sum()
new_infected = np.random.randint(1, INCUBATION_PERIOD, new_infected_count) + day
infected = np.concatenate((infected, new_infected))
infected_lst.append(infected.size)
new_cases_lst.append(new_cases_count)
new_cases_total_lst.append(sum(new_cases_lst))
print(day, infected.size)
plt.figure(figsize=(16, 8))
plt.subplot(311)
plt.title(f"COVID-19 pandemic in {COUNTRY}")
plt.plot(days, new_cases_total_lst)
plt.grid(True)
plt.legend(["Total cases"], loc='upper left')
plt.subplot(312)
plt.bar(days, new_cases_lst, alpha=0.7, color='y')
plt.grid(True)
plt.legend(["New cases"], loc='upper left')
plt.subplot(313)
plt.plot(days, infected_lst, color='r')
plt.grid(True)
plt.legend(["Infected"], loc='upper left')
plt.show()
.
DAYS_OF_SIMULATION - ,
COEF_BASE - ,
COEF_QUARANTINE - ,
DAY_QUARANTINE - ,
INCUBATION_PERIOD - ( ).
, , .
, :
COUNTRY = "Italy"
DAYS_OF_SIMULATION = 366
COEF_BASE = 0.35
COEF_QUARANTINE = 0.135
DAY_QUARANTINE = 74
INCUBATION_PERIOD = 15
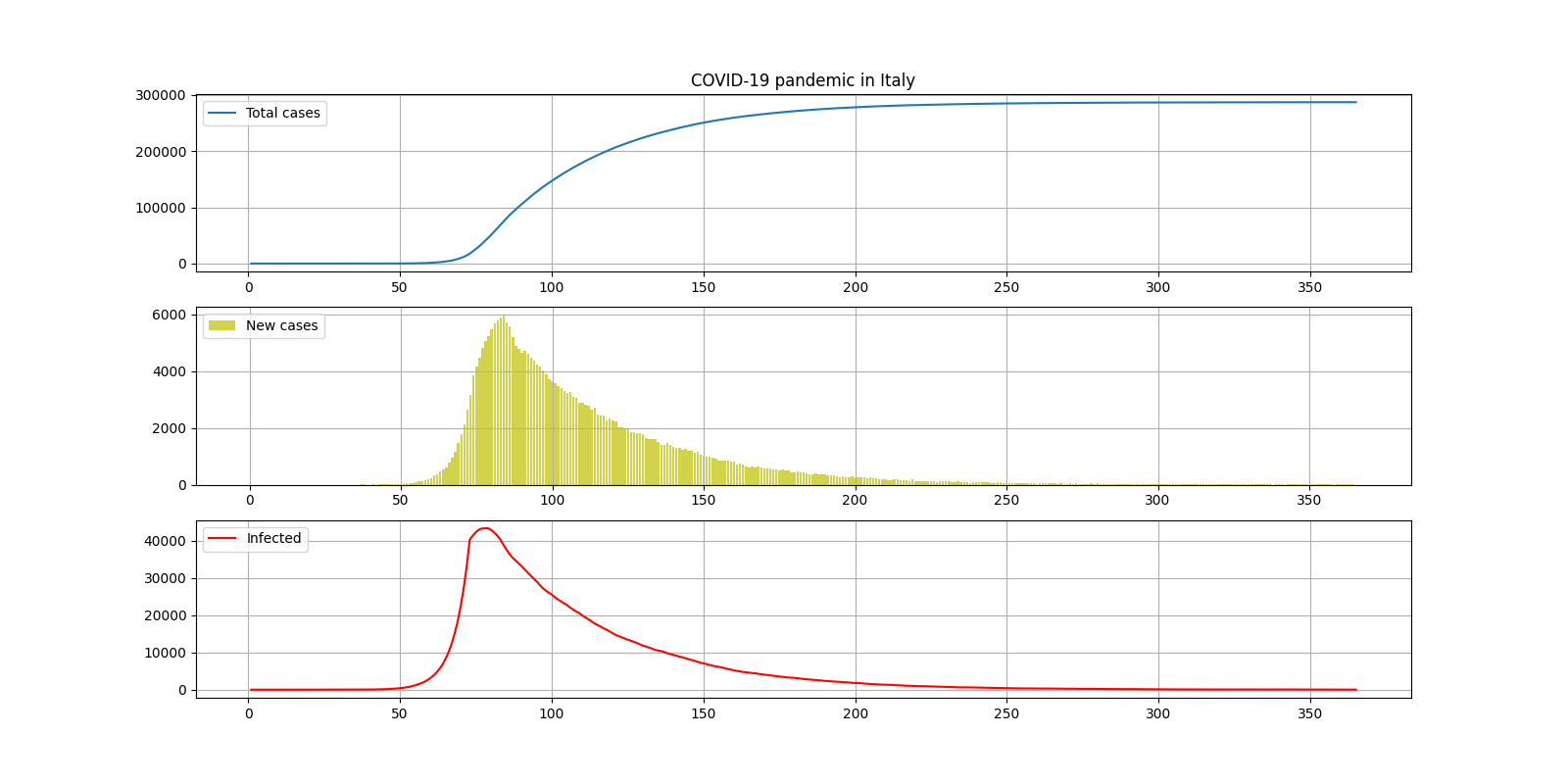
. - , ( 74- ), - , , 14 . , 300000, 250- , 180 ( , , 6 ). , 40000.
:
— 300000
— 6
— 40000
, , :
COUNTRY = "USA"
DAYS_OF_SIMULATION = 366
COEF_BASE = 0.35
COEF_QUARANTINE = 0.135
DAY_QUARANTINE = 83
INCUBATION_PERIOD = 15
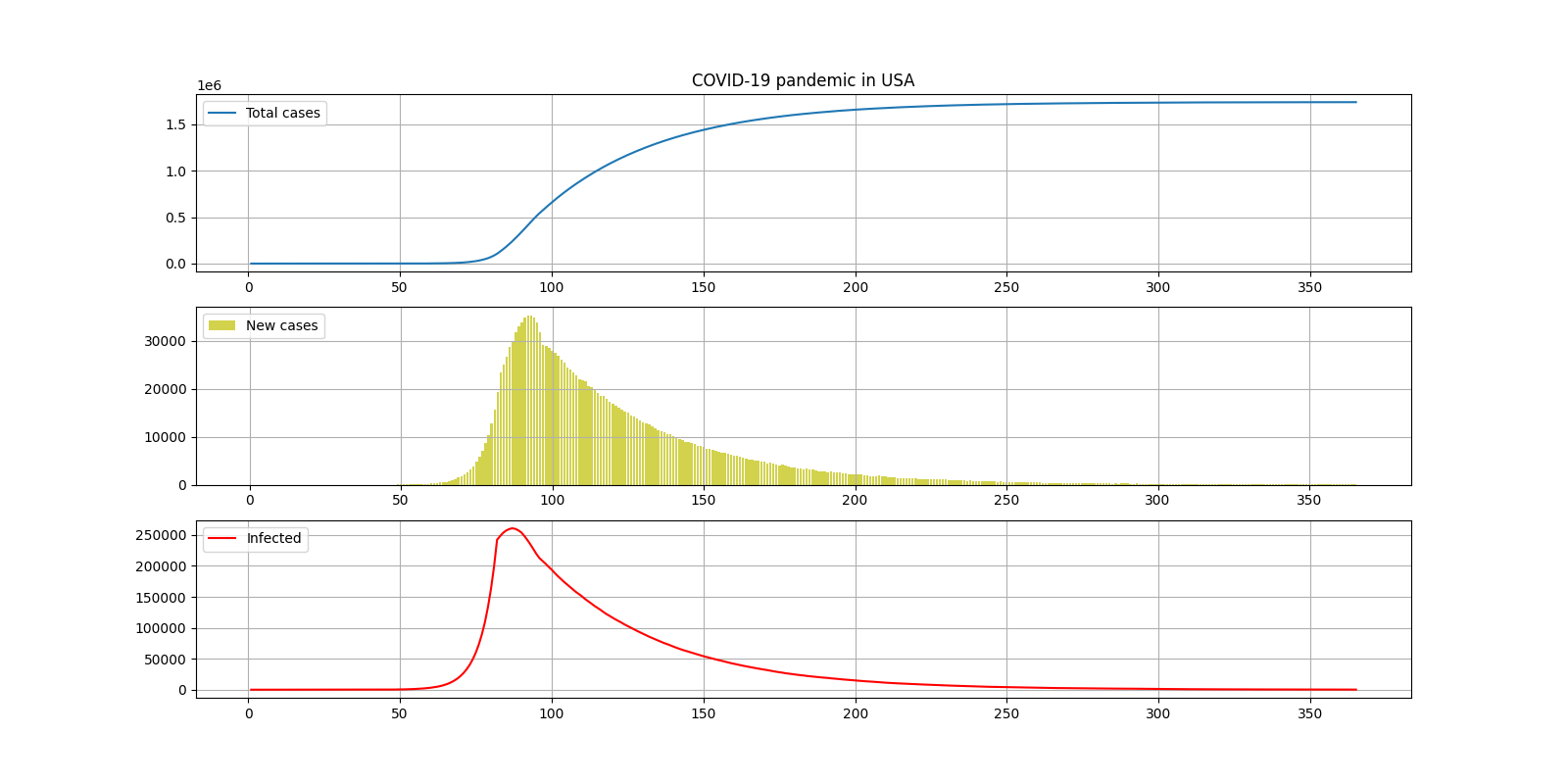
. , , ( DAY_QUARANTINE - ), , COEF_QUARANTINE . .
:
— 1700000
— 6
— 250000
— , . , , . "" . :
- — .
- 1 — .
- 2 — .
.
I.
COUNTRY = "Russia"
DAYS_OF_SIMULATION = 366
COEF_BASE = 0.35
COEF_QUARANTINE = 0.135
DAY_QUARANTINE = 73
INCUBATION_PERIOD = 15
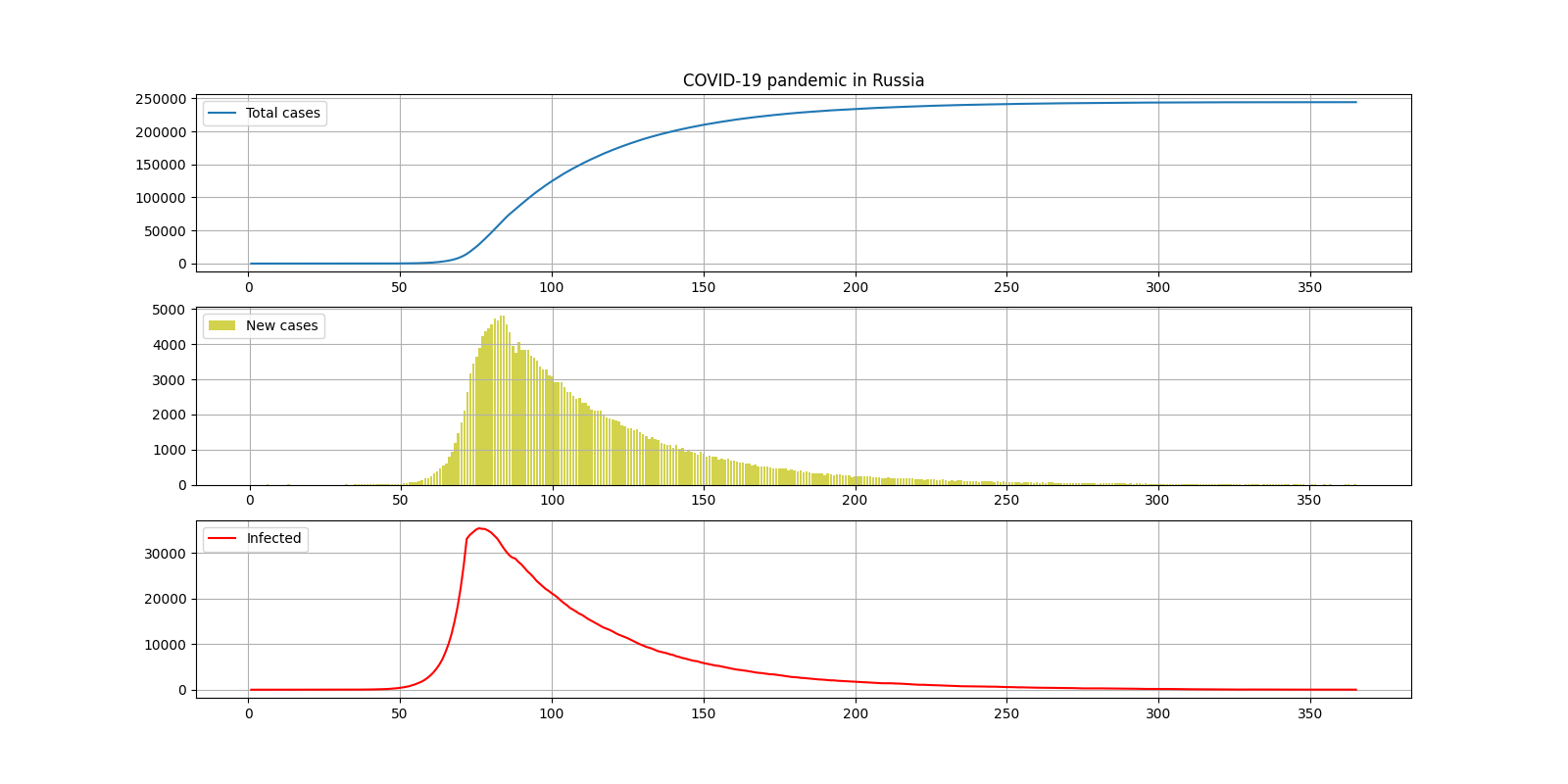
— 250000
— 6
— 35000
II. 1
COUNTRY = "Russia"
DAYS_OF_SIMULATION = 366
COEF_BASE = 0.35
COEF_QUARANTINE = 0.135
DAY_QUARANTINE = 80
INCUBATION_PERIOD = 15
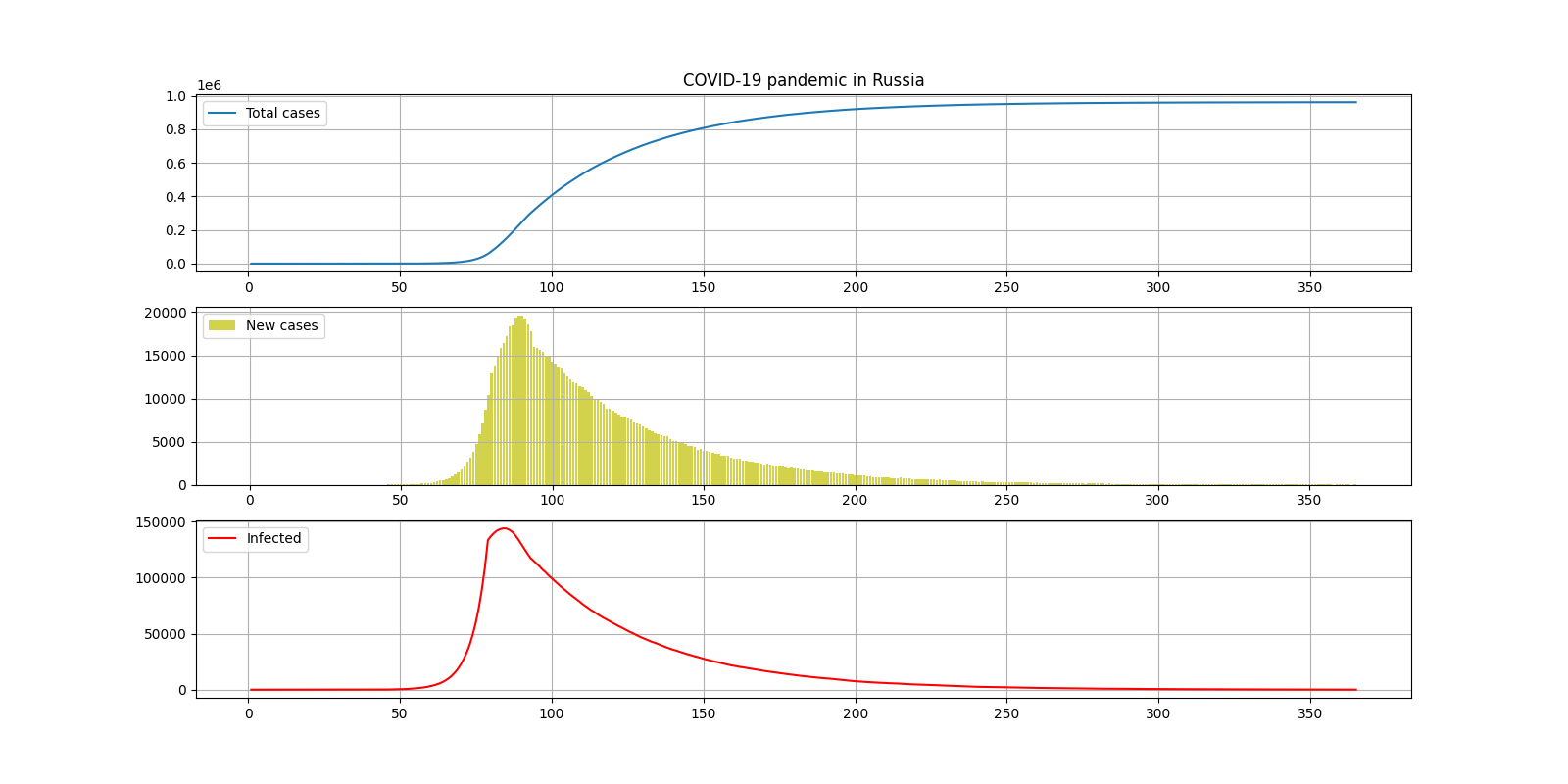
— 950000
— 7
— 140000
III. 2
COUNTRY = "Russia"
DAYS_OF_SIMULATION = 366
COEF_BASE = 0.35
COEF_QUARANTINE = 0.135
DAY_QUARANTINE = 87
INCUBATION_PERIOD = 15
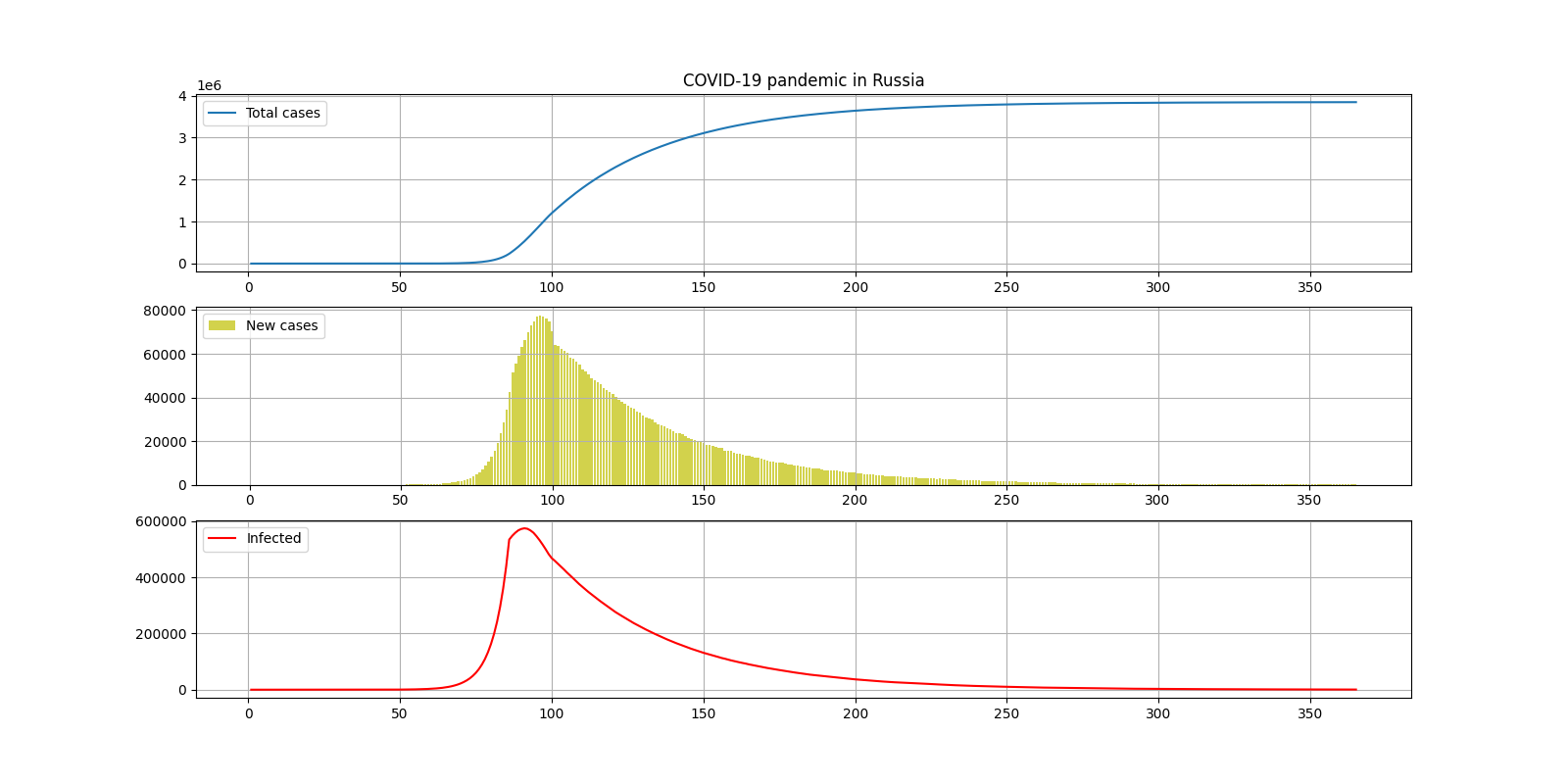
— 4000000
— 8
— 550000
, , -, DAY_QUARANTINE, COEF_QUARANTINE, ( , ). - :
- , ;
- ;
- .
, , . , . , . :
- , , ,
- ,
- ( , ),
- ,
- ,
- … .
I suggest readers to play around with the script, try to simulate the process for other countries, unsubscribe in the comments. It’s also interesting to complicate the simulation by adding more unaccounted factors.