Immune imprinting in childhood: the origin of virus protection
 Almost all of us have heard or read news about the spreading coronavirus. As with any other disease, early diagnosis is important in the fight against the new virus. However, not all infected people show the same set of symptoms, and even scanners at airports designed to detect signs of infection do not always successfully identify a patient among a crowd of passengers. The question arises - why does the same virus manifest itself in different people in different ways? Naturally, the first answer is immunity. However, this is not the only important parameter that affects the variability of symptoms and the severity of the disease. Scientists from the University of California and Arizona (USA) have found that the strength of resistance to viruses depends not only on what subtypes of influenza a person has been ill with over a lifetime, but also on their sequence.What exactly did scientists find out, what methods were used in the study, and how can this work help in the fight against epidemics? We will find answers to these questions in the report of the research group. Go.
Almost all of us have heard or read news about the spreading coronavirus. As with any other disease, early diagnosis is important in the fight against the new virus. However, not all infected people show the same set of symptoms, and even scanners at airports designed to detect signs of infection do not always successfully identify a patient among a crowd of passengers. The question arises - why does the same virus manifest itself in different people in different ways? Naturally, the first answer is immunity. However, this is not the only important parameter that affects the variability of symptoms and the severity of the disease. Scientists from the University of California and Arizona (USA) have found that the strength of resistance to viruses depends not only on what subtypes of influenza a person has been ill with over a lifetime, but also on their sequence.What exactly did scientists find out, what methods were used in the study, and how can this work help in the fight against epidemics? We will find answers to these questions in the report of the research group. Go.Study basis
As we know, in different people the flu manifests itself in different ways. In addition to the human factor (the immune system, the use of antiviral drugs, preventive measures, etc.), the virus itself, or rather its subtype, with which one or another patient is infected, is an important aspect. Each subtype has its own characteristics, including the degree of defeat of various demographic groups. Scientists note that the H1N1 ("swine flu") and H3N2 viruses (Hong Kong influenza), which have become the most common at the moment, affect people of different ages in different ways: H3N2 is the cause of most serious cases of the disease in the elderly, he is also attributed to most deaths ; H1N1 is less deadly, but most often affects middle-aged people and young people.Such differences can be due to both the difference in the rate of evolution of the viruses themselves, and the difference in immune imprinting * in children.Immune imprinting * is a kind of long-term memory of the immune system, formed on the basis of experienced viral attacks on the body and its reactions to them.
In this study, scientists conducted an analysis of epidemiological data to find out whether imprinting in childhood affects the epidemiology of seasonal flu and, if so, does it work mainly through homosubtypic * immune memory or through a wider heterosubtipic * memory.Homosubtypic immunity * - infection with seasonal influenza A viruses contributes to the development of immune defense against a specific subtype of the virus.
Heterosubtypic immunity * - infection with seasonal influenza A viruses promotes the development of immune defense against sub-strains unrelated to this virus.
In other words, children's immunity and everything that he survived leaves his mark on immunity for life. Previous studies have shown that adults have stronger immunity against the types of viruses that they infected in childhood. It has also recently been found that imprinting protects against new subtypes of the avian influenza virus of the same phylogenetic group of hemagglutinin ( hemagglutinin , HA) as during the first infection in childhood.Until recently, narrow cross-protective immunity, specific for variants of one subtype of HA, was considered the main way to protect against seasonal flu. However, there is new evidence that the formation of immunity may be affected by the memory of other influenza antigens (e.g., neuraminidase, NA). Since 1918, three subtypes of HA were recorded among people: H1, H2, and H3. At the same time, H1 and H2 belong to phylogenetic group 1, and H3 to group 2.If we take into account the fact that imprinting most likely causes multiple changes in the immune memory, then we can assume that these changes have a certain hierarchy.Scientists note that since 1977, two subtypes of influenza A, H1N1 and H3N2, have been circulating seasonally among the population. Moreover, the differences in the demographics of infection and in the symptoms were quite obvious, but poorly understood. These differences can be associated with imprinting in childhood: older people were almost certainly exposed to H1N1 in childhood (from 1918 to 1975 it was the only subtype circulating among people). Therefore, at present, these people are better protected from modern seasonal variants of the virus of this subtype. Similarly, in young people, the highest likelihood of imprinting in childhood refers to more modern H3N2 (image No. 1), which is consistent with the relatively low number of clinically recorded cases of H3N2 in this demographic group. Image No. 1: variants of models of the dependence of immunity on imprinting in childhood and the factor of viral evolution.On the other hand, these differences may be related to the evolution of the virus subtypes themselves. So, H3N2 shows a faster drift * of its antigenic phenotype than H1N1.
Image No. 1: variants of models of the dependence of immunity on imprinting in childhood and the factor of viral evolution.On the other hand, these differences may be related to the evolution of the virus subtypes themselves. So, H3N2 shows a faster drift * of its antigenic phenotype than H1N1.Antigen drift * - changes in the immunogenic surface factors of viruses.
For this reason, H3N2 can better avoid previously formed immunity in immunologically experienced (previously ill) adults, while H1N1 can be relatively limited in its effect solely on immunologically inexperienced (previously not sick) children.To test all possible hypotheses, scientists conducted an analysis of epidemiological data, creating likelihood functions for each variant of statistical models, which were compared using the Akaike information criterion (AIC).An additional analysis of the hypothesis was also carried out in which the differences are not due to imprinting in the evolution of viruses.Study preparation
Arizona State Department of Health (ADHS) data was used to model the hypotheses, namely 9,510 cases of seasonal H1N1 and H3N2 across the state. Approximately 76% of the reported cases were recorded in hospitals and laboratories, the remaining cases were not specified in the laboratories. It is also known that approximately half of the laboratory-established cases were quite serious, and therefore led to hospitalization.The data used in the study relate to a period of 22 years: from the flu season 1993–1994 to the season 2014–2015. It is worth noting that the sample size increased sharply after the 2009 pandemic, because this period was excluded from the sample (table No. 1).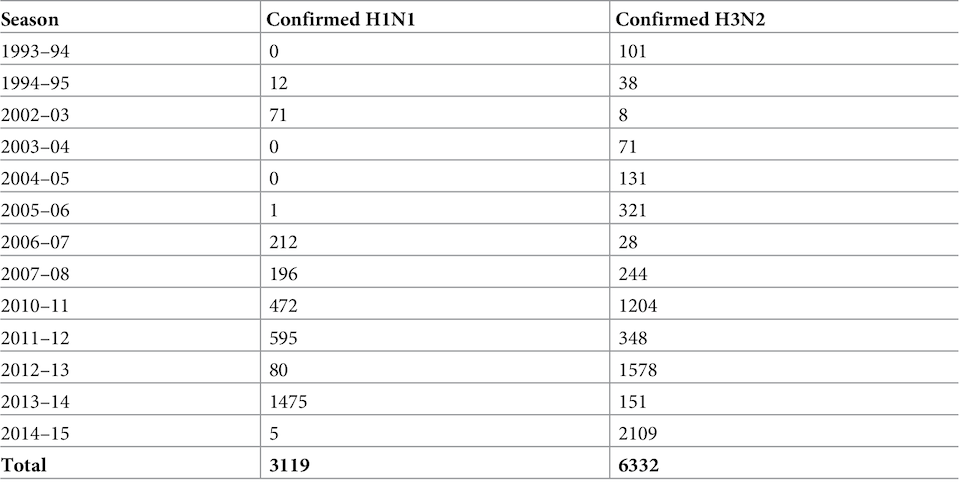 Table 1: Epidemiological data from 1993 to 2015 regarding recorded cases of H1N1 and H3N2 viruses.It is also important to consider that since 2004, US commercial laboratories have been obliged to transmit to the public health authorities all data regarding viral infection of patients. However, most of the analyzed cases (9150/9451) were observed starting from the 2004–2005 season, after this rule came into force.Of all 9,510 cases, 58 were excluded, as these were people with a year of birth before 1918 (their imprinting status cannot be unambiguously determined), and another 1 case due to an incorrectly indicated year of birth. Thus, 9541 cases were included in the analysis model.At the first stage of modeling, the probabilities of imprinting to viruses of H1N1, H2N2 or H3N2 specific for the year of birth were determined. These probabilities reflect the pattern of exposure to influenza A in children and its prevalence over years.Most of the people born between the pandemics in 1918 and 1957 were first infected with the H1N1 subtype. People born between the pandemics of 1957 and 1968, almost all were infected with the H2N2 subtype ( 1A ). And since 1968, the dominant subtype of the virus has been H3N2, which caused the infection of most people from the young demographic group.Despite the prevalence of H3N2, H1N1 has still been circulating seasonally among the population since 1977, causing imprinting in some people born since the mid-1970s (1A ).If imprinting at the level of the HA subtype forms a probability of infection during seasonal flu, then the effect of HA on the H1 or H3 subtypes in early childhood should provide lifelong immunity to more modern variants of the same HA subtype. If imprinting immunity works mostly against certain types of NA (neuraminidase), then lifelong protection will be characteristic of N1 or N2 ( 1B ).If imprinting is based on a broader NA, i.e. Since protection against a wider range of subtypes takes place, people with imprinting from H1 and H2 should be protected from modern seasonal H1N1. At the same time, people with H3 imprinting will be protected only from modern seasonal H3N2 ( 1B ).Scientists note that collinearity (roughly speaking, parallelism) of predictions of various imprinting models ( 1D - 1I ) was inevitable, given the limited variety of antigenic influenza subtypes circulating among the population over the past century.The most important role in the differentiation between imprinting at the level of the HA subtype, NA subtype, or at the level of the HA group is played by middle-aged people who first became infected with H2N2 ( 1B ).Each of the tested models used a linear combination of infection related to age ( 1C ) and infection related to year of birth ( 1D - 1F ) to obtain the distribution of cases of H1N1 or H3N2 ( 1G- 1I ).In total, 4 models were created: the simplest one contained only the age factor, and imprinting factors at the level of the HA subtype, at the level of the NA subtype, or at the level of the HA group were added to more complex models.The curve of the age factor takes the form of a step function in which the relative risk of infection was set to 1 in the 0–4 age group. In addition to the primary age group, there were also the following: 5–10, 11–17, 18–24, 25–31, 32–38, 39–45, 46–52, 53–59, 60–66, 67–73, 74– 80, 81+.In the models that contained the effects of imprinting, it was assumed that the proportion of people in each year of birth with protective imprinting in childhood is proportional to the reduction in the risk of infection.Also, the factor of viral evolution was taken into account in the simulation. For this, data were used that described the annual antigenic progress, which was defined as the average antigenic distance between strains of a particular viral line (H1N1 before 2009, H1N1 after 2009 and H3N2). The "antigenic distance" between two strains of influenza is used as an indicator of the similarity of the antigenic phenotype and potential immune cross-protection.In order to assess the effect of antigenic evolution on the epidemic age distribution, changes in the proportion of cases in children were tested in the seasons when strong antigenic changes occurred.If the level of antigenic drift is a decisive factor in the age-related risk of infection, then the proportion of cases observed in children should be negatively associated with annual antigenic progress. In other words, strains that have not undergone significant antigenic changes compared to the previous season should be unable to avoid pre-existing immunity in adults with immunological experience. Such strains will be more active among the population that does not have immunological experience, that is, among children.
Table 1: Epidemiological data from 1993 to 2015 regarding recorded cases of H1N1 and H3N2 viruses.It is also important to consider that since 2004, US commercial laboratories have been obliged to transmit to the public health authorities all data regarding viral infection of patients. However, most of the analyzed cases (9150/9451) were observed starting from the 2004–2005 season, after this rule came into force.Of all 9,510 cases, 58 were excluded, as these were people with a year of birth before 1918 (their imprinting status cannot be unambiguously determined), and another 1 case due to an incorrectly indicated year of birth. Thus, 9541 cases were included in the analysis model.At the first stage of modeling, the probabilities of imprinting to viruses of H1N1, H2N2 or H3N2 specific for the year of birth were determined. These probabilities reflect the pattern of exposure to influenza A in children and its prevalence over years.Most of the people born between the pandemics in 1918 and 1957 were first infected with the H1N1 subtype. People born between the pandemics of 1957 and 1968, almost all were infected with the H2N2 subtype ( 1A ). And since 1968, the dominant subtype of the virus has been H3N2, which caused the infection of most people from the young demographic group.Despite the prevalence of H3N2, H1N1 has still been circulating seasonally among the population since 1977, causing imprinting in some people born since the mid-1970s (1A ).If imprinting at the level of the HA subtype forms a probability of infection during seasonal flu, then the effect of HA on the H1 or H3 subtypes in early childhood should provide lifelong immunity to more modern variants of the same HA subtype. If imprinting immunity works mostly against certain types of NA (neuraminidase), then lifelong protection will be characteristic of N1 or N2 ( 1B ).If imprinting is based on a broader NA, i.e. Since protection against a wider range of subtypes takes place, people with imprinting from H1 and H2 should be protected from modern seasonal H1N1. At the same time, people with H3 imprinting will be protected only from modern seasonal H3N2 ( 1B ).Scientists note that collinearity (roughly speaking, parallelism) of predictions of various imprinting models ( 1D - 1I ) was inevitable, given the limited variety of antigenic influenza subtypes circulating among the population over the past century.The most important role in the differentiation between imprinting at the level of the HA subtype, NA subtype, or at the level of the HA group is played by middle-aged people who first became infected with H2N2 ( 1B ).Each of the tested models used a linear combination of infection related to age ( 1C ) and infection related to year of birth ( 1D - 1F ) to obtain the distribution of cases of H1N1 or H3N2 ( 1G- 1I ).In total, 4 models were created: the simplest one contained only the age factor, and imprinting factors at the level of the HA subtype, at the level of the NA subtype, or at the level of the HA group were added to more complex models.The curve of the age factor takes the form of a step function in which the relative risk of infection was set to 1 in the 0–4 age group. In addition to the primary age group, there were also the following: 5–10, 11–17, 18–24, 25–31, 32–38, 39–45, 46–52, 53–59, 60–66, 67–73, 74– 80, 81+.In the models that contained the effects of imprinting, it was assumed that the proportion of people in each year of birth with protective imprinting in childhood is proportional to the reduction in the risk of infection.Also, the factor of viral evolution was taken into account in the simulation. For this, data were used that described the annual antigenic progress, which was defined as the average antigenic distance between strains of a particular viral line (H1N1 before 2009, H1N1 after 2009 and H3N2). The "antigenic distance" between two strains of influenza is used as an indicator of the similarity of the antigenic phenotype and potential immune cross-protection.In order to assess the effect of antigenic evolution on the epidemic age distribution, changes in the proportion of cases in children were tested in the seasons when strong antigenic changes occurred.If the level of antigenic drift is a decisive factor in the age-related risk of infection, then the proportion of cases observed in children should be negatively associated with annual antigenic progress. In other words, strains that have not undergone significant antigenic changes compared to the previous season should be unable to avoid pre-existing immunity in adults with immunological experience. Such strains will be more active among the population that does not have immunological experience, that is, among children.Research results
Analysis of data over the years showed that seasonal H3N2 was the main cause of infection among the older population, while H1N1 affected middle-aged and young people (image No. 2).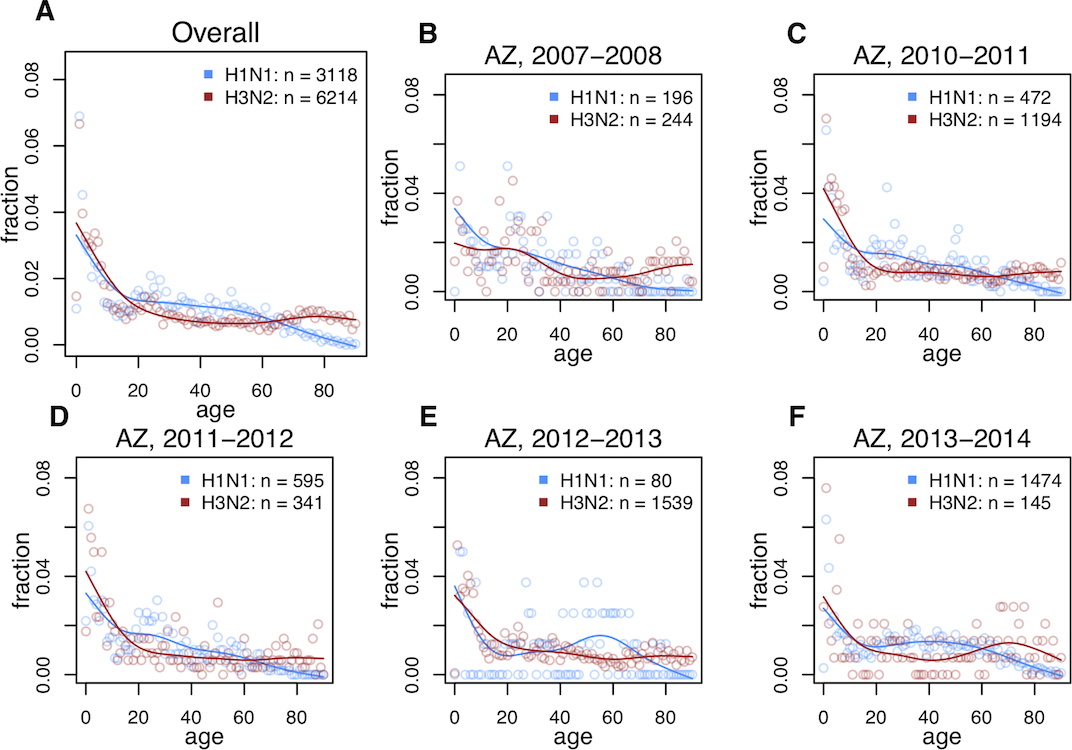 Image No. 2: age distribution of H1N1 and H3N2 flu at different time periods.This pattern was present both in the data before the 2009 pandemic and after it.The data showed that imprinting at the level of the NA subtype prevails over imprinting at the level of the HA subtype (ΔAIC = 34.54). Moreover, imprinting was almost completely absent at the level of the HA group (ΔAIC = 249.06), as was the complete absence of imprinting (ΔAIC = 385.42).
Image No. 2: age distribution of H1N1 and H3N2 flu at different time periods.This pattern was present both in the data before the 2009 pandemic and after it.The data showed that imprinting at the level of the NA subtype prevails over imprinting at the level of the HA subtype (ΔAIC = 34.54). Moreover, imprinting was almost completely absent at the level of the HA group (ΔAIC = 249.06), as was the complete absence of imprinting (ΔAIC = 385.42). Image No. 3: assessment of the conformity of models to the research data.Visual conformity assessment of models ( 3C and3D ) has confirmed that models containing imprinting effects at narrow levels of the NA or HA subtypes provide the best fit for the data used in the study. The fact that a model in which imprinting is absent cannot be supported by data suggests that imprinting is an extremely important aspect of the formation of immunity in the adult population in relation to seasonal influenza subtypes. Nevertheless, imprinting works according to a very narrow specialization, that is, it acts exclusively on a specific subtype, and not on a whole range of influenza subtypes.
Image No. 3: assessment of the conformity of models to the research data.Visual conformity assessment of models ( 3C and3D ) has confirmed that models containing imprinting effects at narrow levels of the NA or HA subtypes provide the best fit for the data used in the study. The fact that a model in which imprinting is absent cannot be supported by data suggests that imprinting is an extremely important aspect of the formation of immunity in the adult population in relation to seasonal influenza subtypes. Nevertheless, imprinting works according to a very narrow specialization, that is, it acts exclusively on a specific subtype, and not on a whole range of influenza subtypes. Table No. 2: assessment of the conformity of models to research data.After taking into account the demographic distribution by age, the expected age-related risk was highest in children and the elderly, which corresponded to the accumulation of immune memory in childhood and a weakening of immune function in the elderly ( 3A shows an approximate curve from the best model). Estimates of imprinting parameters were less than unity, which indicates a slight decrease in relative risk (table No. 2). In the framework of the best model, the estimated decrease in the relative risk from imprinting in childhood was stronger for H1N1 (0.34, 95% CI 0.29–0.42) than for H3N2 (0.71, 95% CI 0.62–0.82).To test the effect of viral evolution on the age distribution of the risk of infection, scientists searched for a decrease in the percentage of infections among children during periods associated with antigenic changes, when strains with high antigenic drift more effectively infected adult immunologically experienced people.An analysis of the data showed a slight negative but insignificant relationship between the annual increase in antigenic activity and the percentage of cases of H3N2 observed in children ( 4A ).
Table No. 2: assessment of the conformity of models to research data.After taking into account the demographic distribution by age, the expected age-related risk was highest in children and the elderly, which corresponded to the accumulation of immune memory in childhood and a weakening of immune function in the elderly ( 3A shows an approximate curve from the best model). Estimates of imprinting parameters were less than unity, which indicates a slight decrease in relative risk (table No. 2). In the framework of the best model, the estimated decrease in the relative risk from imprinting in childhood was stronger for H1N1 (0.34, 95% CI 0.29–0.42) than for H3N2 (0.71, 95% CI 0.62–0.82).To test the effect of viral evolution on the age distribution of the risk of infection, scientists searched for a decrease in the percentage of infections among children during periods associated with antigenic changes, when strains with high antigenic drift more effectively infected adult immunologically experienced people.An analysis of the data showed a slight negative but insignificant relationship between the annual increase in antigenic activity and the percentage of cases of H3N2 observed in children ( 4A ). Image 4: The effect of viral evolution on the age-related risk factor for infection.However, between antigenic changes and the proportion of cases observed in children older than 10 years and in adults, there was no clear relationship. If viral evolution played a major role in this distribution, then as a result, there would be clearer evidence of evolutionary effects among adults, and not just when comparing adults and children under 10 years of age.In addition, if the degree of evolutionary changes in viruses is dominant for subtype-specific differences in the distribution of epidemic age, then when the H1N1 and H3N2 subtypes show the same degree of annual spread of antigen, their age distribution of infections should look more similar.For a more detailed acquaintance with the nuances of the study, I recommend that you look into the report of scientists .
Image 4: The effect of viral evolution on the age-related risk factor for infection.However, between antigenic changes and the proportion of cases observed in children older than 10 years and in adults, there was no clear relationship. If viral evolution played a major role in this distribution, then as a result, there would be clearer evidence of evolutionary effects among adults, and not just when comparing adults and children under 10 years of age.In addition, if the degree of evolutionary changes in viruses is dominant for subtype-specific differences in the distribution of epidemic age, then when the H1N1 and H3N2 subtypes show the same degree of annual spread of antigen, their age distribution of infections should look more similar.For a more detailed acquaintance with the nuances of the study, I recommend that you look into the report of scientists .Epilogue
In this work, scientists analyzed the epidemiological data of cases of infection with H1N1, H3N2 and H2N2. Data analysis showed a clear dependence of imprinting in childhood and the degree of risk of infection in adulthood. In other words, if a child was infected in the 1950s when H1N1 was circulating, and H3N2 was absent, then in adulthood the probability of H3N2 infection will be much more than the probability of catching H1N1.The main conclusion of this study is that it is important not only what a person was sick in childhood, but also in what sequence. Immune memory, which is formed throughout life, actively "records" the data of the first viral infections, which contributes to a more effective response to them in adulthood.Scientists hope that their work will make it possible to better predict which age groups are more susceptible to the effects of a particular influenza subtype. This knowledge can help prevent the spread of epidemics, especially if you need to distribute a limited number of vaccines among the population.This study is not aimed at finding super-cures for any type of flu, although that would be great. It aims at what is much more real and important at the moment - preventing the spread of infection. If we cannot instantly get rid of the virus, then we must have all the possible tools to contain it. One of the most faithful allies of any epidemic is the negligent attitude towards it both on the part of the state as a whole, and each person in particular. Panic, of course, is not needed, because it can only make it worse, but safety precautions will never hurt.Thank you for your attention, remain curious, take care of yourself and your loved ones and have a great weekend for everyone, guys! :)A bit of advertising :)
Thank you for staying with us. Do you like our articles? Want to see more interesting materials? Support us by placing an order or recommending to your friends, cloud VPS for developers from $ 4.99 , a unique analog of entry-level servers that was invented by us for you: The whole truth about VPS (KVM) E5-2697 v3 (6 Cores) 10GB DDR4 480GB SSD 1Gbps from $ 19 or how to divide the server? (options are available with RAID1 and RAID10, up to 24 cores and up to 40GB DDR4).Dell R730xd 2 times cheaper at the Equinix Tier IV data center in Amsterdam? Only we have 2 x Intel TetraDeca-Core Xeon 2x E5-2697v3 2.6GHz 14C 64GB DDR4 4x960GB SSD 1Gbps 100 TV from $ 199 in the Netherlands!Dell R420 - 2x E5-2430 2.2Ghz 6C 128GB DDR3 2x960GB SSD 1Gbps 100TB - from $ 99! Read about How to Build Infrastructure Bldg. class c using Dell R730xd E5-2650 v4 servers costing 9,000 euros per penny?Source: https://habr.com/ru/post/undefined/
All Articles