يعد اكتشاف الشذوذ مهمة مهمة للتعلم الآلي. لا توجد طريقة محددة لحلها ، لأن كل مجموعة بيانات لها خصائصها الخاصة. ولكن في نفس الوقت ، هناك العديد من الأساليب التي تساعد على النجاح. أريد أن أتحدث عن أحد هذه الأساليب - ترميز السيارات.
أي مجموعة بيانات تختار؟
القضية الأكثر إلحاحًا في حياة أي عالم بيانات. لتبسيط القصة ، سأستخدم مجموعة بيانات بسيطة في الهيكل ، والتي سننشئها هنا.
import os
import numpy as np
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
from matplotlib.colors import Normalize
def gen_normal_distribution(mu, sigma, size, range=(0, 1), max_val=1):
bins = np.linspace(*range, size)
result = 1 / (sigma * np.sqrt(2*np.pi)) * np.exp(-(bins - mu)**2 / (2*sigma**2))
cur_max_val = result.max()
k = max_val / cur_max_val
result *= k
return result
خذ بعين الاعتبار مثال دالة. قم بإنشاء توزيع عادي مع μ = 0.3 و σ = 0.05:
dist = gen_normal_distribution(0.3, 0.05, 256, max_val=1)
print(dist.max())
>>> 1.0
plt.plot(np.linspace(0, 1, 256), dist)
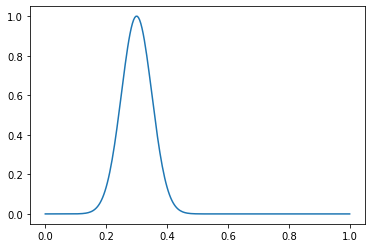
أعلن معلمات مجموعة البيانات لدينا:
in_distribution_size = 2000
out_distribution_size = 200
val_size = 100
sample_size = 256
random_generator = np.random.RandomState(seed=42)
ووظائف توليد الأمثلة طبيعية وغير طبيعية. تعتبر التوزيعات بحد أقصى عادي وغير طبيعي - باثنين:
def generate_in_samples(size, sample_size):
global random_generator
in_samples = np.zeros((size, sample_size))
in_mus = random_generator.uniform(0.1, 0.9, size)
in_sigmas = random_generator.uniform(0.05, 0.5, size)
for i in range(size):
in_samples[i] = gen_normal_distribution(in_mus[i], in_sigmas[i], sample_size, max_val=1)
return in_samples
def generate_out_samples(size, sample_size):
global random_generator
out_samples = generate_in_samples(size, sample_size)
out_additional_mus = random_generator.uniform(0.1, 0.9, size)
out_additional_sigmas = random_generator.uniform(0.01, 0.05, size)
for i in range(size):
anomaly = gen_normal_distribution(out_additional_mus[i], out_additional_sigmas[i], sample_size, max_val=0.12)
out_samples[i] += anomaly
return out_samples
هذا ما يبدو عليه المثال العادي:
in_samples = generate_in_samples(in_distribution_size, sample_size)
plt.plot(np.linspace(0, 1, sample_size), in_samples[42])

وغير طبيعي - مثل هذا:
out_samples = generate_out_samples(out_distribution_size, sample_size)
plt.plot(np.linspace(0, 1, sample_size), out_samples[42])
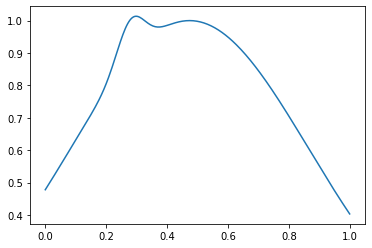
إنشاء صفائف مع العلامات والعلامات:
x = np.concatenate((in_samples, out_samples))
y = np.concatenate((np.zeros(in_distribution_size), np.ones(out_distribution_size)))
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, shuffle=True, random_state=42)
. , , 2 1 (). , ( ), :
x_val_out = generate_out_samples(val_size, sample_size)
x_val_in = generate_in_samples(val_size, sample_size)
x_val = np.concatenate((x_val_out, x_val_in))
y_val = np.concatenate((np.ones(val_size), np.zeros(val_size)))
, , Sklearn: SVM . , , .
from sklearn.metrics import classification_report
from sklearn.metrics import f1_score
One class SVM
from sklearn.svm import OneClassSVM
OneClassSVM nu — .
out_dist_part = out_distribution_size / (out_distribution_size + in_distribution_size)
svm = OneClassSVM(nu=out_dist_part)
svm.fit(x_train, y_train)
>>> OneClassSVM(cache_size=200, coef0=0.0, degree=3, gamma='scale', kernel='rbf',
max_iter=-1, nu=0.09090909090909091, shrinking=True, tol=0.001,
verbose=False)
:
svm_prediction = svm.predict(x_val)
svm_prediction[svm_prediction == 1] = 0
svm_prediction[svm_prediction == -1] = 1
sklearn — classification_report, Anomaly detection , precision recall, :
print(classification_report(y_val, svm_prediction))
>>> precision recall f1-score support
0.0 0.57 0.93 0.70 100
1.0 0.81 0.29 0.43 100
accuracy 0.61 200
macro avg 0.69 0.61 0.57 200
weighted avg 0.69 0.61 0.57 200
, . f1-score , .
Isolation forest
, , - ?
from sklearn.ensemble import IsolationForest
, . :
out_dist_part = out_distribution_size / (out_distribution_size + in_distribution_size)
iso_forest = IsolationForest(n_estimators=100, contamination=out_dist_part, max_features=100, n_jobs=-1)
iso_forest.fit(x_train)
>>> IsolationForest(behaviour='deprecated', bootstrap=False,
contamination=0.09090909090909091, max_features=100,
max_samples='auto', n_estimators=100, n_jobs=-1,
random_state=None, verbose=0, warm_start=False)
Classification report? — Classification report!
iso_forest_prediction = iso_forest.predict(x_val)
iso_forest_prediction[iso_forest_prediction == 1] = 0
iso_forest_prediction[iso_forest_prediction == -1] = 1
print(classification_report(y_val, iso_forest_prediction))
>>> precision recall f1-score support
0.0 0.50 0.91 0.65 100
1.0 0.53 0.10 0.17 100
accuracy 0.51 200
macro avg 0.51 0.51 0.41 200
weighted avg 0.51 0.51 0.41 200
RandomForestClassifier
, - " ?" , :
from sklearn.ensemble import RandomForestClassifier
random_forest = RandomForestClassifier(n_estimators=100, max_features=100, n_jobs=-1)
random_forest.fit(x_train, y_train)
>>> RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=None, max_features=100,
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=100,
n_jobs=-1, oob_score=False, random_state=None, verbose=0,
warm_start=False)
random_forest_prediction = random_forest.predict(x_val)
print(classification_report(y_val, random_forest_prediction))
>>> precision recall f1-score support
0.0 0.57 0.99 0.72 100
1.0 0.96 0.25 0.40 100
accuracy 0.62 200
macro avg 0.77 0.62 0.56 200
weighted avg 0.77 0.62 0.56 200
Autoencoder
, : . .
, "" , . 2 : Encoder' Decoder', .
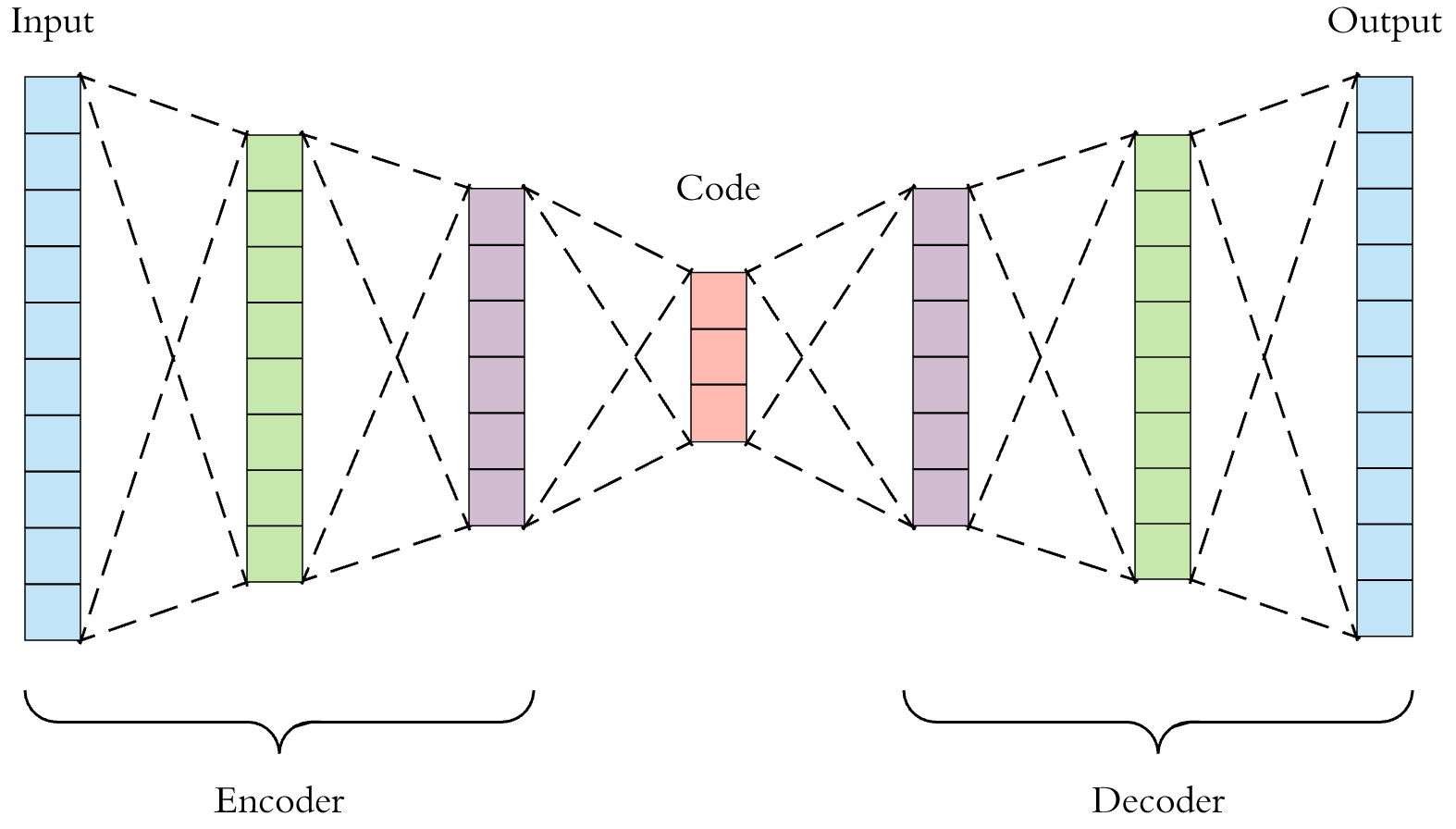
"" , , .
? , , , . , 9% 91% . , . , , : .
.
PyTorch, :
import torch
from torch import nn
from torch.utils.data import TensorDataset, DataLoader
from torch.optim import Adam
:
batch_size = 32
lr = 1e-3
. , , "" . , ( , learning rate, ) , , .
train_in_distribution = x_train[y_train == 0]
train_in_distribution = torch.tensor(train_in_distribution.astype(np.float32))
train_in_dataset = TensorDataset(train_in_distribution)
train_in_loader = DataLoader(train_in_dataset, batch_size=batch_size, shuffle=True)
test_dataset = TensorDataset(
torch.tensor(x_test.astype(np.float32)),
torch.tensor(y_test.astype(np.long))
)
test_loader = DataLoader(test_dataset, batch_size=batch_size, shuffle=False)
val_dataset = TensorDataset(torch.tensor(x_val.astype(np.float32)))
val_loader = DataLoader(val_dataset, batch_size=batch_size, shuffle=False)
. 4 , ( 2 : μ, — σ, ; 4 , ).
class Autoencoder(nn.Module):
def __init__(self, input_size):
super(Autoencoder, self).__init__()
self.encoder = nn.Sequential(
nn.Linear(input_size, 128),
nn.LeakyReLU(0.2),
nn.Linear(128, 64),
nn.LeakyReLU(0.2),
nn.Linear(64, 16),
nn.LeakyReLU(0.2),
nn.Linear(16, 4),
nn.LeakyReLU(0.2),
)
self.decoder = nn.Sequential(
nn.Linear(4, 16),
nn.LeakyReLU(0.2),
nn.Linear(16, 64),
nn.LeakyReLU(0.2),
nn.Linear(64, 128),
nn.LeakyReLU(0.2),
nn.Linear(128, 256),
nn.LeakyReLU(0.2),
)
def forward(self, x):
x = self.encoder(x)
x = self.decoder(x)
return x
model = Autoencoder(sample_size).cuda()
criterion = nn.MSELoss()
per_sample_criterion = nn.MSELoss(reduction="none")
optimizer = Adam(model.parameters(), lr=lr, weight_decay=1e-5)
- loss' , , boxplot' :
def save_score_distribution(model, data_loader, criterion, save_to, figsize=(8, 6)):
losses = []
labels = []
for (x_batch, y_batch) in data_loader:
x_batch = x_batch.cuda()
output = model(x_batch)
loss = criterion(output, x_batch)
loss = torch.mean(loss, dim=1)
loss = loss.detach().cpu().numpy().flatten()
losses.append(loss)
labels.append(y_batch.detach().cpu().numpy().flatten())
losses = np.concatenate(losses)
labels = np.concatenate(labels)
losses_0 = losses[labels == 0]
losses_1 = losses[labels == 1]
fig, ax = plt.subplots(1, figsize=figsize)
ax.boxplot([losses_0, losses_1])
ax.set_xticklabels(['normal', 'anomaly'])
plt.savefig(save_to)
plt.close(fig)
:
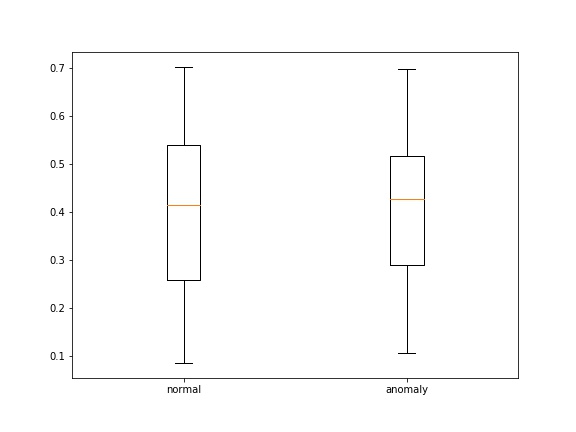
:
experiment_path = "ood_detection"
!rm -rf $experiment_path
os.makedirs(experiment_path, exist_ok=True)
epochs = 100
for epoch in range(epochs):
running_loss = 0
for (x_batch, ) in train_in_loader:
x_batch = x_batch.cuda()
output = model(x_batch)
loss = criterion(output, x_batch)
optimizer.zero_grad()
loss.backward()
optimizer.step()
running_loss += loss.item()
print("epoch [{}/{}], train loss:{:.4f}".format(epoch+1, epochs, running_loss))
plot_path = os.path.join(experiment_path, "{}.jpg".format(epoch+1))
save_score_distribution(model, test_loader, per_sample_criterion, plot_path)
>>>
epoch [1/100], train loss:8.5728
epoch [2/100], train loss:4.2405
epoch [3/100], train loss:4.0852
epoch [4/100], train loss:1.7578
epoch [5/100], train loss:0.8543
...
epoch [96/100], train loss:0.0147
epoch [97/100], train loss:0.0154
epoch [98/100], train loss:0.0117
epoch [99/100], train loss:0.0105
epoch [100/100], train loss:0.0097

50

100
, . , :
def get_prediction(model, x):
global batch_size
dataset = TensorDataset(torch.tensor(x.astype(np.float32)))
data_loader = DataLoader(dataset, batch_size=batch_size, shuffle=False)
predictions = []
for batch in data_loader:
x_batch = batch[0].cuda()
pred = model(x_batch)
predictions.append(pred.detach().cpu().numpy())
predictions = np.concatenate(predictions)
return predictions
def compare_data(xs, sample_num, data_range=(0, 1), labels=None):
fig, axes = plt.subplots(len(xs))
sample_size = len(xs[0][sample_num])
for i in range(len(xs)):
axes[i].plot(np.linspace(*data_range, sample_size), xs[i][sample_num])
if labels:
for i, label in enumerate(labels):
axes[i].set_ylabel(label)
:
x_test_pred = get_prediction(model, x_test)
compare_data([x_test[y_test == 0], x_test_pred[y_test == 0]], 10, labels=["real", "encoded"])
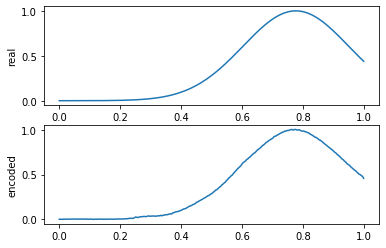
:

X? .
Difference score
— . , , , . .
def get_difference_score(model, x):
global batch_size
dataset = TensorDataset(torch.tensor(x.astype(np.float32)))
data_loader = DataLoader(dataset, batch_size=batch_size, shuffle=False)
predictions = []
for (x_batch, ) in data_loader:
x_batch = x_batch.cuda()
preds = model(x_batch)
predictions.append(preds.detach().cpu().numpy())
predictions = np.concatenate(predictions)
return (x - predictions)
from sklearn.ensemble import RandomForestClassifier
test_score = get_difference_score(model, x_test)
score_forest = RandomForestClassifier(max_features=100)
score_forest.fit(test_score, y_test)
>>> RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=None, max_features=100,
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=100,
n_jobs=None, oob_score=False, random_state=None,
verbose=0, warm_start=False)
, : 2 — . , 2 : , — difference_score, — . , , .
? . difference_score , ( ) , , . , , difference_score , ( ). .
:
val_score = get_difference_score(model, x_val)
prediction = score_forest.predict(val_score)
print(classification_report(y_val, prediction))
>>> precision recall f1-score support
0.0 0.76 1.00 0.87 100
1.0 1.00 0.69 0.82 100
accuracy 0.84 200
macro avg 0.88 0.84 0.84 200
weighted avg 0.88 0.84 0.84 200
. :
indices = np.arange(len(prediction))
wrong_indices = indices[(prediction == 0) & (y_val == 1)]
x_val_pred = get_prediction(model, x_val)
compare_data([x_val, x_val_pred], wrong_indices[0])
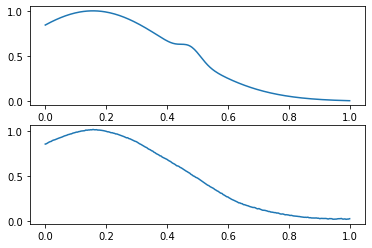
? :
plt.imshow(val_score[wrong_indices], norm=Normalize(0, 1, clip=True))

:
plt.imshow(val_score[(prediction == 1) & (y_val == 1)], norm=Normalize(0, 1, clip=True))

:
plt.imshow(val_score[(prediction == 0) & (y_val == 0)], norm=Normalize(0, 1, clip=True))

, : .
Difference histograms
. . — , "", .
, difference score
print("test score: [{}; {}]".format(test_score.min(), test_score.max()))
>>> test score: [-0.2260764424351479; 0.26339245919832344]
, :
def score_to_histograms(scores, bins=10, data_range=(-0.3, 0.3)):
result_histograms = np.zeros((len(scores), bins))
for i in range(len(scores)):
hist, bins = np.histogram(scores[i], bins=bins, range=data_range)
result_histograms[i] = hist
return result_histograms
test_histogram = score_to_histograms(test_score, bins=10, data_range=(-0.3, 0.3))
val_histogram = score_to_histograms(val_score, bins=10, data_range=(-0.3, 0.3))
plt.title("normal histogram")
plt.bar(np.linspace(-0.3, 0.3, 10), test_histogram[y_test == 0][0])

plt.title("anomaly histogram")
plt.bar(np.linspace(-0.3, 0.3, 10), test_histogram[y_test == 1][0])
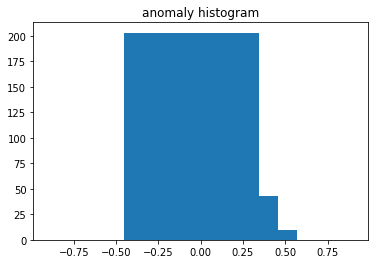
, "", .
histogram_forest = RandomForestClassifier(n_estimators=10)
histogram_forest.fit(test_histogram, y_test)
>>> RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=None, max_features='auto',
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=10,
n_jobs=None, oob_score=False, random_state=None,
verbose=0, warm_start=False)
val_prediction = histogram_forest.predict(val_histogram)
print(classification_report(y_val, val_prediction))
>>> precision recall f1-score support
0.0 0.83 0.99 0.90 100
1.0 0.99 0.80 0.88 100
accuracy 0.90 200
macro avg 0.91 0.90 0.89 200
weighted avg 0.91 0.90 0.89 200
— . , , , , . () .
— . ? VAE, ( 4 ) . . CVAE, - . , , , ..
GAN', (), . ( ).
, , .
Statistical parametric
- GMM — Gaussian mixture modelling + Akaike or Bayesian Information Criterion
- HMM — Hidden Markov models
- MRF — Markov random fields
- CRF — conditional random fields
Robust statistic
- minimum volume estimation
- PCA
- estimation maximisation (EM) + deterministic annealing
- K-means
Non-parametric statistics
- histogram analysis with density estimation on KNN
- local kernel models (Parzen windowing)
- vector of feature matching with similarity distance (between train and test)
- wavelets + MMRF
- histogram-based measures features
- texture features
- shape features
- features from VGG-16
- HOG
Neural networks
- self organisation maps (SOM) or Kohonen's
- Radial Basis Functions (RBF) (Minhas, 2005)
- LearningVector Quantisation (LVQ)
- ProbabilisticNeural Networks (PNN)
- Hopfieldnetworks
- SupportVector Machines (SVM)
- AdaptiveResonance Theory (ART)
- Relevance vector machine (RVM)
, Data science' . Computer vision, . ( — !) — FARADAY Lab. — , .
c:
